BioTechnology: Principles And Processes
Principles Of Biotechnology
- Biotechnology can be broadly defined as “using living organisms or their products for commercial purposes.”
- As such, biotechnology has been practiced by human society since the beginning of recorded history in activities such as baking bread, brewing alcoholic beverages, and breeding food crops or domestic animals.
- A narrower and more specific definition of biotechnology is “the commercial application of living organisms or their products, which involves deliberate manipulation of their DNA molecules.”
- This definition implies a set of laboratory techniques developed within the last 20 years that have been responsible for the tremendous scientific and commercial interest in biotechnology.
- Some other available definitions of biotechnology are as follows:
- “The application of biological organisms, systems, or processes to manufacturing and service industries.” British Biotechnologist
- “The integrated use of biochemistry, microbiology, and genetic engineering sciences in order to achieve technological (industrial) application of capabilities of microorganisms, cultured tissue cells, and parts thereof.”-European Federation of Biotechnology
- “Controlled use of biological agents such as microorganisms and cellular components for beneficial use.” US National Science Foundation
- The development of biotechnology can be studied considering its growth that occurred in two phases: (a) Traditional (old) biotechnology and (b) new (modern) biotechnology.
Read and Learn More NEET Biology Notes
Traditional Biotechnology
- Traditional biotechnology includes processes that are based on the natural capabilities of microorganisms.
- It is also called conventional technology; it has been used for many centuries.
- Curd; vinegar; ghee; wine, beer, and other alcoholic beverages; and idli, dosa, cheese, paneer, and some other foods have been produced using traditional bio-technology.
- In Indian Ayurveda, the production of asva, arista, etc., is done through traditional biotechnology.
- According to some people, traditional biotechnology is, therefore, an art rather than a science.
Modern Biotechnology
- When extremely new and useful traits in crop varieties and animal breeds are created with the help of genetic engineering, it is called modern biotechnology.
- It was developed in 1970.
- For example, in vitro fertilization leading to a “test tube baby,” synthesizing a gene and using it, developing a DNA vaccine, or correcting a defective gene are all parts of modern biotechnology.
- Among many, the two main techniques that gave birth to modern biotechnology are as follows:
- Genetic engineering: The techniques which change the chemistry of genetic material (DNA and RNA) to introduce these into host organisms and, thus, alter the phenotype of the host organism are called genetic engineering (recombinant DNA technology).
- To maintain microbial contamination-free (sterile) surrounding in chemical engineering: Due to such type of maintenance, only desired microorganisms/cells will be formed in large number for the manufacture of biotechnological products such as antibiotics, vaccines, enzymes, hormones, and blood clotting factors. It is essential to have complete aseptic conditions.
Concept of Genetic Engineering
- Combining DNA from different existing organisms such as plants, animals, and bacteria results in modified organisms with a combination of traits from the parents.
- This sharing of DNA information occurs naturally through sexual reproduction and has been exploited in plant and animal breeding for a number of years.
- However, sexual reproduction (recombination) can occur between the individuals of same species. Genetic engineering is the manipulation of prokaryotic as well as eukaryotic DNA.
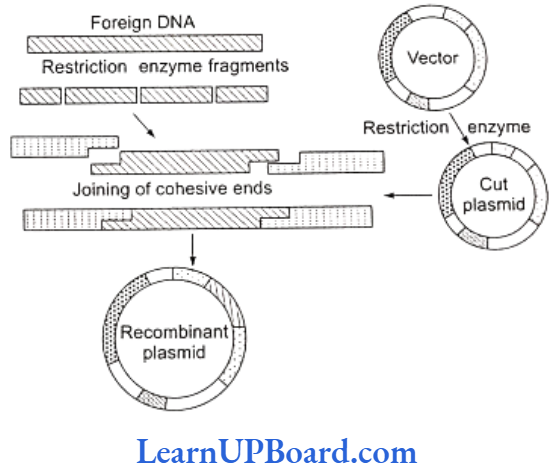
- It involves the breakage of a DNA molecule at two de- sired places to isolate a specific DNA segment which is then inserted in another DNA molecule at a desired position.
- The product, thus, obtained is called recombinant DNA and the technique is often called genetic engineering.
- The cutting of the DNA at specific locations became possible by so called “molecular scissors,” i.e., restriction enzymes.
- In chromosomes, there is a specific “ori” site or the origin of replication which initiates the replication.
- Therefore, for the duplication of any foreign DNA in an organism, it should be linked with the ori sites so that the foreign DNA can duplicate and multiply within the organism.
- This is called gene cloning. Multiple copies of any template DNA can be produced by using gene cloning.
- The construction of the first recombinant DNA emerged from the possibility of linking a gene encoding antibiotic resistance with a native plasmid (autonomously replicating circular extra-chromosomal DNA) of Salmonella typhimurium.
- Stanley Cohen and Herbert Boyer accomplished this in 1972 by isolating the antibiotic resistance gene by cutting out a piece of DNA from a plasmid which was responsible for conferring antibiotic resistance.
- These plasmid DNA molecules act as vectors to transfer the piece of DNA attached to it.
- As mosquito acts as an insect vector to transfer the malarial parasite into human body, in the same way, a plasmid can be used as a vector to deliver an alien piece of DNA into the host organism.
- The linking of antibiotic resistance gene with the plasmid vector became possible with enzyme DNA ligase, which acts on cut DNA molecules and joins their ends.
- This makes a new combination of circular autonomously replicating DNA created in vitro known as recombinant DNA.
- When this DNA is transferred into Escherichia coli, a bacterium closely related to Salmonella, it replicates using the new host’s DNA polymerase enzyme and makes multiple copies.
- The ability to multiply copies of antibiotic resistance gene in E. coli was called cloning of antibiotic resistance gene in E. coli.
- Therefore, there are three basic steps in genetically modifying an organism:
- Identification of DNA with desirable genes.
- Introduction of the identified DNA into the host.
- Maintenance of introduced DNA in the host and transfer of the DNA to its progeny.
” biological molecules”
Tools Of Recombinant DNA Technology
The technology or genetic engineering involves restriction enzymes, ligase enzymes, polymerase enzymes, vectors, and the host organism.
Restriction Enzymes
- In the late 1960’s, scientists Stewart Linn and Werner Arber isolated samples of two types of enzymes responsible for phage growth restriction in E. coli bacteria.
- One of these enzymes was methylated DNA while the other was cleaved unmethylated DNA at a wide variety of locations along the length of the molecule. The first type of enzyme was called methylase while the other was called restriction nuclease.
- These enzymatic tools were important for scientists who were gathering the tools needed to “cut and paste” DNA molecules.
- What was needed now was a tool that would cut DNA at specific sites, rather than at random sites along the length of the molecule, so that scientists can cut DNA- molecules in a predictable and reproducible way.
Site-Specific Nuclease
- This important development came when H.O. Smith, K.W. Wilcox, and T.J. Kelley isolated and characterized the first restriction nuclease whose functioning depended on a specific DNA nucleotide sequence.
- Working with Haemophilus influenzae bacteria, this group isolated an enzyme called Hind II that always cut DNA molecules at a particular point within a spe- cific sequence of six base pairs.
- This sequence is
- 5′ GT (pyrimidine: T or C) (purine: A or G) AC 3′
- 3′ CA (purine: A or G) (pyrimidine: T or C) TG 5′
- They found that Hind II enzyme always cuts directly in the center of this sequence.
- Wherever this particular sequence of six base pairs occurs unmodified in a DNA molecule, Hind II will cleave both DNA strands or backbones between the third and the fourth base pairs of the sequence.
- Moreover, Hind II will cleave both DNA strands or backbones between the third and the fourth base pairs of the sequence will only cleave a DNA molecule at this particular site. For this reason, this specific base sequence is known as the “recognition sequence” for Hind II.
- Hind II is just one example of the class of enzymes known as restriction nucleases.
- In fact, more than 900 restriction enzymes, some sequences specific and some not, have been isolated from over 230 strains of bacteria since the initial discovery of Hind II.
- These restriction enzymes generally have names that reflect their origin.
- The first letter (in italics) of the name comes from the genus and the second two letters (in italics) come from the species of the prokaryotic cell from which they were isolated.
- Next is the strain of the organism and last is the Roman numeral indicating the order of discovery (the order in which the enzymes were isolated from single strains of bacteria).
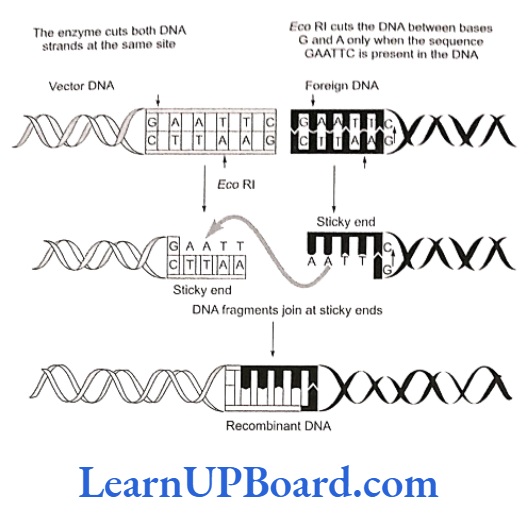
- For example, Eco RI comes from Escherichia coli RY strain (and was the first endonuclease isolated from bacteria) while Hind II comes from Haemophilus in- fluenzae strain Rd.
- Nucleases are further described by the addition of the prefix “endo” or “exo” to the name. The term “endonuclease” applies to sequence-specific nucleases that break nucleic acid chains somewhere in the interior, rather than at the ends of the molecule.
- Nucleases that function by removing nucleotides from the ends of the molecules are called exonucleases.
- Three main classes of restriction endonucleases: type 1, type 2, and type 3, have been described, each distinguished by a slightly different mode of action.
- Out of these three types, type 1 and type 3 restriction enzymes are not used in recombinant DNA technology.
- Type 2 restriction enzymes are used in recombinant DNA technology, because they can be used in vitro to recognize and cut within the specific DNA sequence typically consisting of 4-8 nucleotides.
- Type 1 enzymes recognize specific sites within the DNA but do not cut at these sites.
- Hence, heterogeneous population of DNA fragments is produced. Therefore, type 1 enzymes do not take part in the technology. Type 3 enzymes recognize a specific sequence of DNA molecule. Thus, the products of type 3 enzymes are homogeneous population of DNA fragments; so, they cannot be used for genetic engineering experiments.
- The DNA segments cut by restriction enzymes are palindromic, i.e., the nucleotide sequences of these DNA pieces read the same both backwards and forward when the orientation of reading is kept same, e.g., madam.
- Blunt or flush ends are produced by many restriction enzymes which cleave both strands of DNA at exactly the same nucleotide position-in the center of recognition site. For example, Small recognizes six-nucleotide palindromic sequence.
- It cuts both DNA strands producing blunt ends.
- Sticky or cohesive ends are produced when restriction enzymes do not cut DNA at the same nucleotide position but cut the recognition sequence unequally. This produces short, single-stranded overhangs at each end. These are known as sticky ends. For example, Eco RI recognizes 6-nucleotide palindromic sequence.
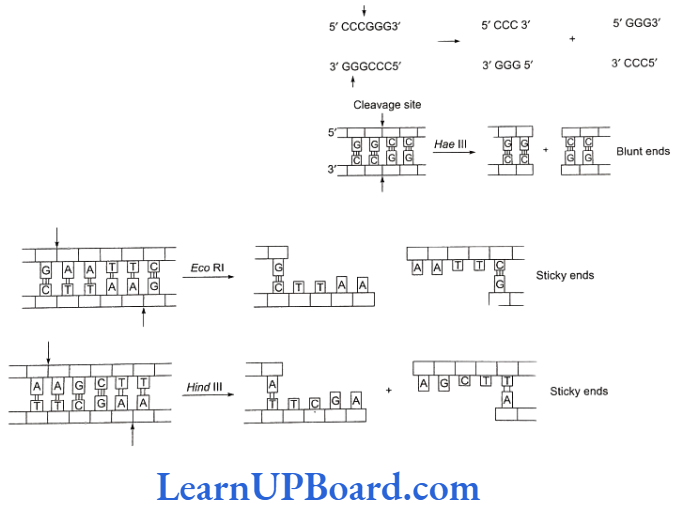
- This restriction endonuclease cuts both DNA strands unequally, producing 5′ overhangs of four nucleotides.
- The stickiness helps enzyme ligase to make the DNA pieces join.
” biomolecules structure”
Other Enzymes Used in Recombinant DNA Technology
- In addition to restriction enzymes, there are several other enzymes that play an important role in DNA technology.
- Three of the important ones are DNA ligase, alkaline phosphatase, and DNA polymerase.
- DNA ligase: This enzyme forms phosphodiester bonds between adjacent nucleotides and covalently links two individual fragments of double-stranded DNA. The action of ligase enzyme requires a phosphate group at the 5′ carbon of one nucleotide and a hydroxyl group at the 3′ carbon of the adjacent nucleotide to form phosphodiester bond between these two nucleotides. The enzyme used most often in the rDNA technology is T4 DNA ligase, which is encoded by phage T4.
- Alkaline phosphatase (AP): As mentioned above, ligation absolutely requires the presence of 5′ phosphate group at the DNA site to be ligated. If this phosphate group is removed, this DNA can- not be ligated. The enzyme alkaline phosphatase is used to remove the phosphate group from the 5′ end of a DNA molecule, leaving a free 5′ hy- droxyl group. This enzyme can be isolated from bacteria (BAP) or calf intestine (CAP). It is used to prevent unwanted self-ligation of vector DNA molecules in the procedures of rDNA technology.
However, the ligation of the vector to the insert can occur as the insert still has its 5′ phosphate. - DNA polymerase: DNA polymerase I (DNA Pol I) enzyme polymerizes DNA synthesis on DNA template or complementary DNA (cDNA). It also catalyzes 5′ 3′ and 3′ 5′ exonucleolytic degradation of DNA. The other two enzymes are DNA Pol II and DNA Pol III. These have almost similar catalytic activity. DNA Pol III is about several times more active than the other two. Where there is preformed DNA template, it pro- duces a parallel strand in the presence of ATP.
Separation and Isolation of DNA Fragments
- After the cutting of DNA by restriction enzymes, fragments of DNA are formed.
- These fragments can be separated by a technique called gel electrophoresis.
- Electrophoresis is a technique of separation of charged molecules under the influence of an electrical field so that they migrate in the direction of the electrode bearing opposite charge, i.e., positively charged molecules move towards cathode (-ve electrode) and negatively charged molecules travel towards anode (+ve electrode), through a medium/matrix.
- This technique was developed by A. Tiselius in 1937.
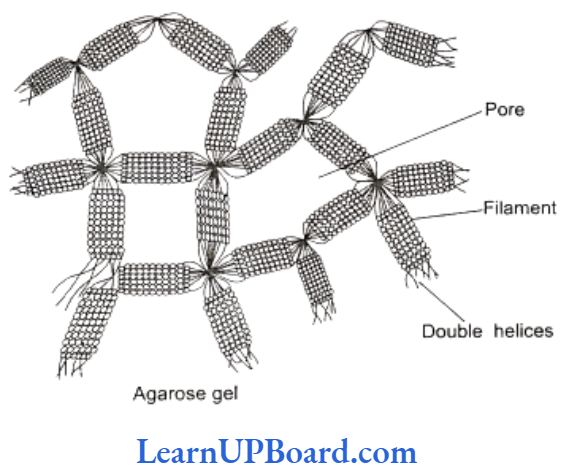
- Nowadays, the most commonly used matrix is agarose which is a polysaccharide extracted from sea weeds.
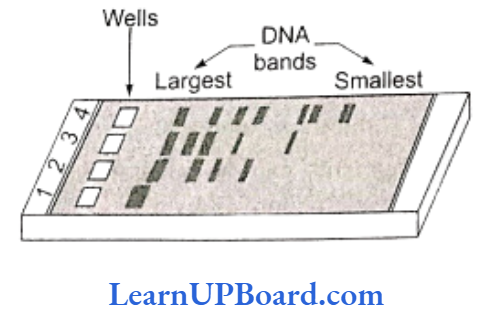
- DNA fragments separate according to size through the pores of agarose gel.
- Hence, the smaller the fragment size, the farther it moves.
- Agarose dissolves in hot water. When this solution is cooled, double helices form. These become arranged laterally and produce thick filaments.
- These filaments become cross-linked to form the gel.
- Pore size depends on agarose concentration.
- Separated DNA fragments can be seen only after staining the DNA with a compound ethidium bromide followed by exposure to UV radiations as bright orange colored bands. The separated bands of DNA are cut out from the agarose gel and extracted from the gel piece. This step is called elution. Several techniques are used for eluting the DNA from the gel piece. These purified DNA fragments are used in the formation of recombinant DNA by linking them with cloning vectors.
- Cloning vectors: Another important tool for genetic engineering is the vehicle for cloning, called vector. A vector carries a foreign DNA sequence into a given host cell. Bacterial plasmids and bacteriophages are considered the most useful. This is because of the following reasons:
- These are independent of the control of chromosomal DNA.
- Bacteriophage genomes occur in very large numbers in bacterial cells.
- The copies of plasmids per cell range from only a few to hundred or even more.
- Certain essential features should be present in a DNA molecule to act as a cloning vector.
- Origin of replication (ori): This is a DNA sequence which serves as a starting point for replication. When a DNA fragment gets associated with ori, foreign DNA into the vector would also replicate inside the host cell. Some vectors possess origin which favors the formation of high copy numbers and, hence, are preferred.
- Selectable marker: A vector should also include a selectable marker. This is a gene which would permit the selection of host cells containing vector from amongst those which do not possess vector. Common selectable markers include genes encoding antibiotic resistance such as ampicillin resistance or enzymes such as ẞ-galactosidase (product of lac Z gene of lac operon). These genes can be identified by a color reaction.
- Recognition sites: A vector should possess a unique restriction site that would allow a particular enzyme to cut the vector only once. This site would be recognized by the commonly used restriction enzymes. If there are more than one recognition sites in a vector, several fragments would be produced. Generally, the vectors used possess unique recognition sites for several restriction enzymes in a small region of DNA. This is known as polylinker or multiple cloning site (MCS). Such cloning site offers a choice of restriction enzymes. Unique restriction endonuclease recognition site enables the insertion of foreign DNA into the vector for the production of recombinant DNA. The foreign DNA is inserted and made to join (ligate) at a specific restriction site, generally, in antibiotic resistance gene.
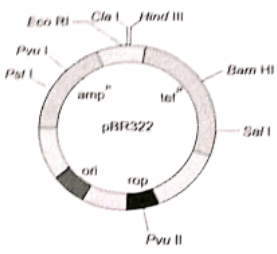
- pBR322 has genes for resistance against two antibiotics (tetracycline and ampicillin). An origin of replication and a variety of restriction sites for cloning of restriction fragments are obtained through cleavage with a specific enzyme. Foreign DNA is inserted at a site located in one of the two genes for resistance against antibiotics, so that it will inactivate one of the two resistance genes.
- The insert bearing plasmids can be selected by their ability to grow in a medium containing only one of the two antibiotics and their failure to grow in a medium containing both antibiotics. The plasmids carrying no insert, on the other hand, will be able to grow in a media containing one or both antibiotics. In this way, the presence of resistance genes against ampicillin and tetracycline allows the selection of Escherichia coli colonies transformed with plasmids carrying the desired foreign cloned DNA fragment.
- Size of the vector: The cloning vector should be small in size. Large molecules have a tendency to break down during purification. These are also difficult to manipulate.
Different Types Of Vectors
Several types of vectors satisfying the above characters have been developed. The following are some of the commonly used vectors.
Plasmids
- These are extra-chromosomal, non-essential self-replicating, usually circular, and double-stranded DNA molecules occurring in some bacteria and also a few yeasts.
- Some of the characters carried by plasmids may not be required for normal bacterial metabolism but may be of great advantage, e.g., antibiotic resistance.
- pBR322 is one of the standard cloning vectors widely used in gene cloning experiments.
- This vector has been restructured by inserting genes for antibiotic resistance.
- It is named after Boliver and Rodriguez who prepared this vector.
” types of biomolecules “
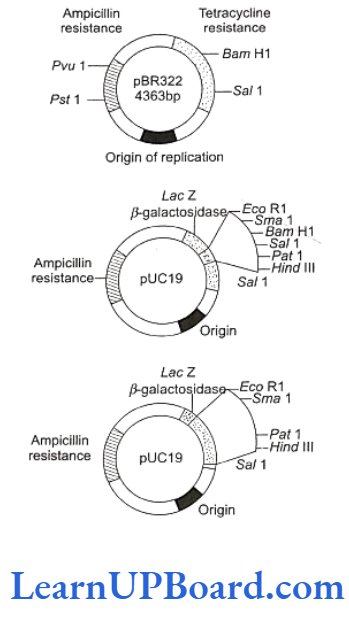
- PUC (named after the University of California) is another such reconstructed plasmid vector.
- The vectors mentioned above are able to replicate only in E. coli.
- Therefore, many vectors constructed for eukaryotic cells are also functional in E. coli.
- These vectors are called shuttle vectors.
- The vectors contain two types of origin of replication and selectable marker genes one for the eukaryotic cell and the other for E. coli.
- The common example of this type is yeast episomal plasmid (Yep).
- In plants, tumor-inducing (Ti) plasmid of bacterium Agrobacterium tumfaciens has been modified to function as a vector.
Vectors Based on Bacteriophages
Bacteriophages are viruses which infect bacterial cells, produce new phages inside the host bacterium, and are released from the host cell to again infect other bacterial cells. M13 and lambda (2) phages are in common use.
Cosmids
Cosmids combine some features of plasmid and cos (cohesive end sites) of phage lambda (cosmid = cos + plasmid).
Yeast Artificial Chromosome Vectors
Yeast artificial chromosome (YAC) contains telomeric sequence, centromere, and autonomously replicating sequence from yeast chromosomes. It also has suitable restriction enzyme sites and genes useful as selectable markers.
Bacterial Artificial Chromosome Vectors
- Bacterial artificial chromosome (BAC) is based on the F-plasmid (fertility) of E. coli. It contains genes for the replication and maintenance of F-factor, selectable marker, and cloning sites.
- Color reaction: Due to the inactivation of antibiotics, the selection of recombinants becomes a burdensome process because it requires simultaneous plating on two plates having different antibiotics. Thus, an alternative selectable marker is developed to differentiate recombinants and non-recombinants on the basis of their ability to produce color in the presence of a chromogenic substance. Now, a recombinant DNA is inserted in the coding sequence of enzyme ẞ-galac-tosidase. This causes the inactivation of the enzyme; it is called insertional inactivation. If the plasmid in the bacterium does not have an insert, the presence of a chromogenic substrate gives blue-colored colonies. The presence of insert results in the insertional inac- tivation of B-galactosidase. Therefore, the colonies do not produce any color. These colonies are marked as recombinant colonies.
Vectors for Cloning Genes in Plants and Animals
- We know the procedure of transferring genes into plants and animals from bacteria and viruses.
- The procedure to transfer genes to transform eukaryotic cells and force them to do what the bacteria or viruses require is also known.
- For example, Agrobacterium tumifaciens, a pathogen (disease causing agent) of several dicot plants, is able to transfer a piece of DNA known as T-DNA to convert normal plant cells into tumor and direct these tumor cells to secrete the chemicals required by the pathogen. Similarly, retroviruses (cause leukosis or sarcoma types of cancer) in animals including humans are able to change normal cells into cancerous cells.
- The Ti plasmid of Agrobacterium tumifaciens has been modified into a cloning vector which is not pathogenic to plants. However, it is still able to use the procedure to deliver genes of our interest into various plants.
- Similarly, retroviruses are used to carry desirable genes into animal cells.
- Thus, once a gene or DNA fragment is joined to a suitable vector, it is transferred into a bacterial plant or animal host where it undergoes multiplication.
Host Cell
- Competent host cell is required for transformation with recombinant DNA.
- After the formation of recombinant DNA, propagation of it must occur inside a living system or a host.
- Different types of available host cells are E. coli, yeast, and animal and plant cells.
- The type of host cell to be used depends on the aim of cloning experiment.
- Eukaryotic cells will be the preferred host for the expression of some eukaryotic proteins.
- Yeast cells are preferred because these are the simplest eukaryotic organisms and, like bacteria, are single celled, genetically well characterized, and easy to grow and manipulate.
- Plant and animal cells can be used for protein expression either in tissue culture or as cells in the whole organism to create genetically modified (GM) crops and animals.
- As DNA is a hydrophilic molecule, it cannot pass through cell membrane.
- Therefore, the bacterial cells should be capable of up- taking DNA.
- This is accomplished by treating them with specific concentration of a divalent cation, e.g., Ca2+, making them competent which causes an efficient entry of DNA into the bacterium through the pores in its cell wall.
- Recombinant DNA can be forced into such cells by incubating the cells with recombinant DNA on ice, followed by placing them briefly at 42°C (heat shock), and then putting them back on ice. As a result, bacteria get enabled to pick up recombinant DNA.
- There are other methods to introduce foreign DNA into host cells. These are briefly described in the following subsections.
Microinjection
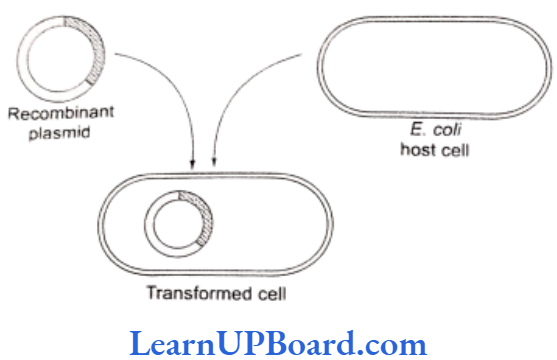
- In the microinjection method, the recombinant DNA is directly injected into the nucleus of animal cell by using micro-needles or micro-pipettes. It is used in oocytes, eggs, and embryo. Jeffey S. Chamberlain et. al. (1993) of Human Genome Center, Michigan University, USA, have cured mice that inherited a neuromuscular disease which is like the muscular dystrophy of humans.
Direct DNA Injection
Direct injection of DNA into skeletal muscle led to the possibility of using gene as vaccines. Due to low level of expression, therapeutic benefits for the treatment of genetic disorder could not be derived. This method gave birth to the concept of DNA vaccine or genetic immunization.
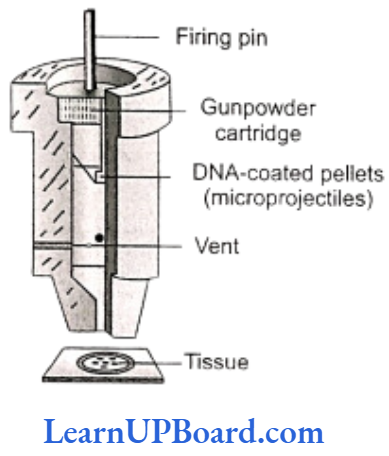
Gene Gun or Biolistics
New technologies such as gene gun are also available for vector-less direct gene transfer. DNA coated onto microscopic pellets is literally shot into target cells. Although it is developed for plants, it is also used for animal cells for promoting tissue repair or reducing healing time. This method made great impact in the field of vaccine development.
Process Of Recombinant DNA Technology
Recombinant technology is a complicated process. Several steps lead to the desired goal. The major steps are as follows:
- Isolation of DNA
- Digestion of DNA by restriction endonuclease enzyme
- Gene amplification
- Introduction of recombinant DNA into host cells.
- Identification of recombinants
- Gene product manufacture
- Processing
” introduction to biomolecules “
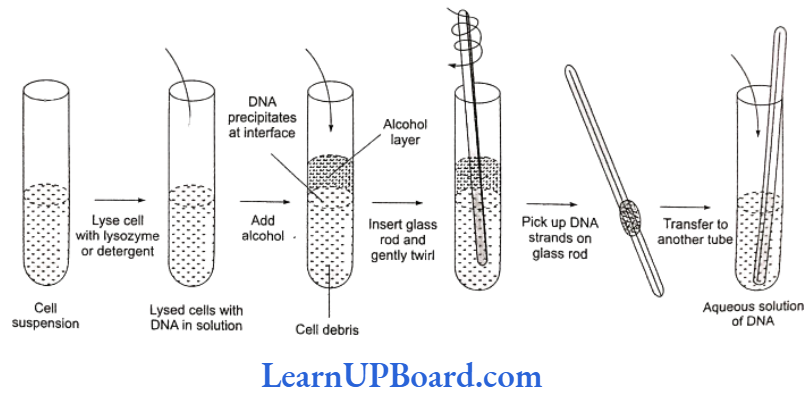
Isolation of DNA: Isolation of the Genetic Material (DNA)
- Nucleic acid (DNA or RNA) is the genetic material of all organisms. It is DNA in majority of organisms.
- For cutting the DNA with restriction enzymes, it needs to be pure and free from other macromolecules.
- Because DNA is covered with membranes, it has to break the cell open to release DNA and other macro-molecules such as RNA, proteins, polysaccharides, and lipids.
- It is obtained by treating the bacterial cells/plant or animal tissues with enzymes such as lysozyme (bacteria), cellulase (plant cells), and chitinase (fungus).
- As we know that genes are present on long molecules of DNA intertwined with proteins such as histones, RNA can be removed by treating with ribonuclease while proteins can be removed by treating with protease.
- Other molecules are removed by proper treatments. Purified DNA finally precipitates out after the addition of chilled ethanol.
- This is seen as a collection of fine threads in suspension.
DNA Digestion by Restriction Enzymes
- The vector and the target DNA fragment can be separately digested with the same restriction enzyme.
- The digested vector and the target DNA fragment are then incubated together in the presence of DNA ligase enzyme.
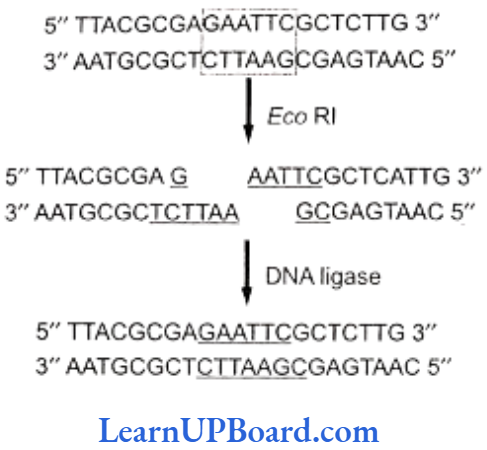
- Incubation results in bonding of two types of DNA by phosphodiester bonds between them.
- Thus, the deoxyribose-phosphate backbones of vector molecule and the target DNA fragment are covalently linked, forming a recombinant DNA molecule.
- Another possibility in this experiment is the rejoining of the sticky ends of the vector molecule itself, forming a circular vector DNA molecule that is without foreign DNA molecule.
- This possibility is eliminated by treating digested vector with alkaline phosphatase or by using different restriction enzymes.
biomolecules protein
Gene Amplification
- Gene amplification is the process of selective multiplication of a specific region of DNA molecule.
- The process has also been used to produce DNA fragments for cloning.
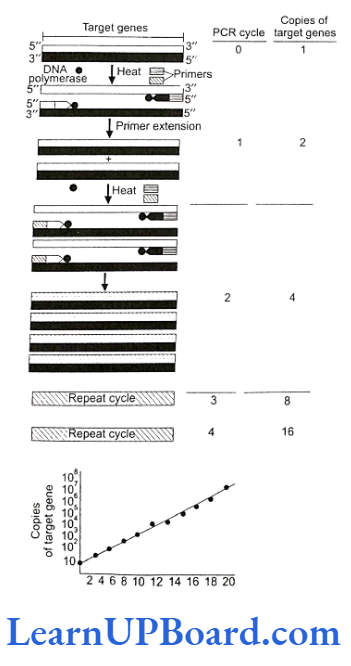
- Amplification is achieved by a special method known as polymerase chain reaction (PCR) developed by Kary Mullis in 1985 for which he shared Nobel Prize in 1993.
- The principle underlying the technique is to heat double-stranded DNA molecule to a high temperature so that the two DNA strands separate into single-stranded DNA molecules.
- If these single-stranded molecules are copied by a DNA polymerase, it would lead to the duplication of the original DNA molecule; if these events are repeated many times, then multiple copies of the original DNA sequence can be generated.
- The basic requirements of a PCR reaction are as follows:
- DNA template: Any source that contains one or more target DNA molecules to be amplified can be taken as a template.
- Primers: Primers, which are oligo-nucleotides, usually 10-18 nucleotides long, hybridize to the target DNA region, one to each strand of the double helix. Two primers are required and these primers are oriented with their ends facing each other, allowing the synthesis of the DNA towards one another.
- Enzyme: DNA polymerase which is stable at high temperatures (>90°) is required to carry out the synthesis of new DNA. The polymerase which is generally used in PCR reactions is Taq polymerase (isolated from bacterium Thermus aquaticus). Other thermostable polymerases can also be used.
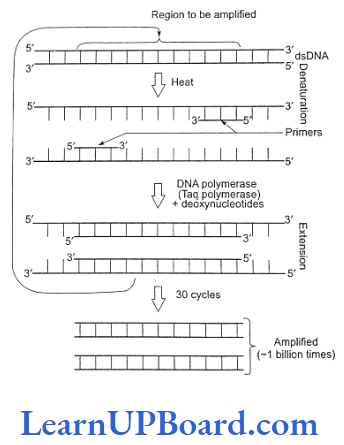
Working Mechanism of PCR
- A single PCR amplification cycle involves three basic steps: denaturation, annealing, and extension (polymerization).
- Denaturation: In the denaturation step, the tar- get DNA is heated to a high temperature (usually 94°C), resulting in the separation of the two strands. Each single strand of the target DNA then acts as a template for DNA synthesis.
- Annealing: In this step, the two oligo-nucleotide primers anneal (hybridize) to each of the single- stranded template DNA since the sequence of primers is complementary to the 3′ ends of template DNA. This step is carried out at a lower temperature depending on the length and sequence of the primers.
- Primer extension (polymerization): The final step is extension, wherein Taq DNA polymerase (of a thermophilic bacterium Thermus acquaticus) synthesizes the DNA region between the primers, using dNTPs (deoxynucleoside triphosphates) and Mg2+. It means the primers are extended towards each other so that the DNA segment lying between the two primers is copied. The optimum temperature for this polymerization step is 72°C. To begin the second cycle, the DNA is again heated to convert all newly synthesized DNA into single strands, each of which can now serve as a template for the synthesis of more new DNA. Thus, the extension product of one cycle can serve as a template for subsequent cycles and each cycle essentially doubles the amount of DNA from the previous cycle. As a result, from a single template molecule, it is possible to generate 2″ molecules after n number of cycles.
Application of PCR
Some of the areas of application of PCR are briefly mentioned here.
- Diagnosis of pathogens: Pathologists use techniques based on detecting specific enzymes or antibodies against disease-related proteins. But these techniques cannot be used for detecting infectious agents that are difficult to culture or that persist at very low levels in infected cells. To overcome these problems, PCR-based assays have been developed that detect the presence of gene sequences of infectious agents.
- Diagnosis of specific mutation: PCR can be used to detect the presence of a specific mutation that is r sponsible for causing a particular genetic disease be- fore the actual onset of the disease. By using PCR, phenylketonuria, muscular dystrophy, sickle-cell anemia, AIDS, hepatitis, Chlamydia, and tuberculosis can be diagnosed.
- DNA fingerprinting: PCR is of immense value in generating abundant amount of DNA for analysis in the DNA fingerprinting technique used in forensic science to link a suspect’s DNA to the DNA recovered at a crime scene.
- Detection of specific microorganisms: PCR is also used for detecting specific microorganisms from the environment samples of soil, sediments, and water.
- In prenatal diagnosis: It is useful to detect a genetic disease in fetus before birth. If the disease is not curable, abortion is recommended.
- Diagnosis of plant pathogens: Many diseases of plants can be detected by using PCR. For example, viroids (associated with apple, grape, citrus, pear, etc.), viruses (such as TMV and bean yellow mosaic virus), bacteria, mycoplasmas, etc.
- In paleontology: PCR is used to clone the DNA fragments from the mummified remains of humans and extinct animals such as wooly mammoth and dinosaurs.
Introduction of Recombinant DNA into Host Cells
- Once the recombinant DNA molecule has been multiplied, it needs to be inserted into host cells.
- Many methods for introduction are available.
- The selection of a method depends upon the type of vector and the host cell along with other things.
- Some common methods are as follows:
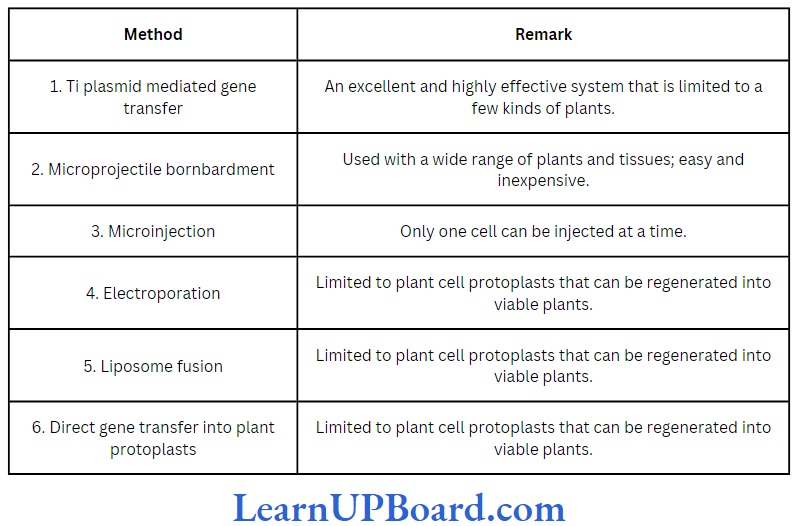
- Transformation: This is a method where cells take up DNA from their surroundings. Since many cells such as those of E. coli, yeast, and mammals do not naturally absorb DNA, they need to be made competent. Mandel and Higa (1970) observed that E. coli cells can be made competent to take up external DNA by suspending them in cold calcium chloride.
” carbohydrates biomolecules”
- Transfection: In this method, DNA is mixed with charged substances such as calcium phosphate and cationic liposomes. These are spread on the recipient host cells. Calcium ions carry foreign DNA and release it inside the cell since calcium gets precipitated in the form of calcium phosphate, thus, transferring the DNA by endocytosis.
- Microinjection and macroinjection: Specially designed micromanipulator is used to inject DNA into cytoplasm or the nucleus of a recipient cell or protoplast. The method is used for the direct introduction of DNA into plant or animal cells without using special eukaryotic vectors.
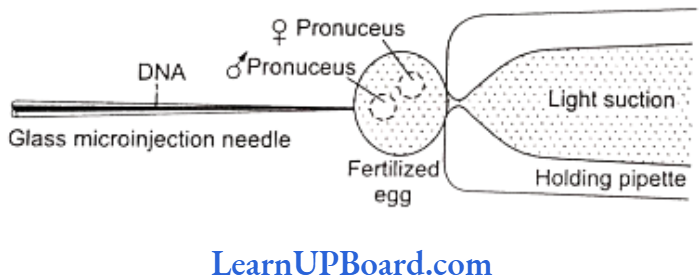
- Microprojection (biolistics or particle gun): Tungsten or gold particles (microparticles) coated with DNA are accelerated to a very high initial velocity. These microprojectiles are carried by other (nylon) microprojectiles or the bullet, causing them to penetrate the cell walls of intact target cells or tissues.
- Electroporation: Short electrical impulses of high field strength are given. These increase the permeability of protoplast membrane by creating transient microscopic pores, thus, making the entry of DNA molecules into the cells much easier.
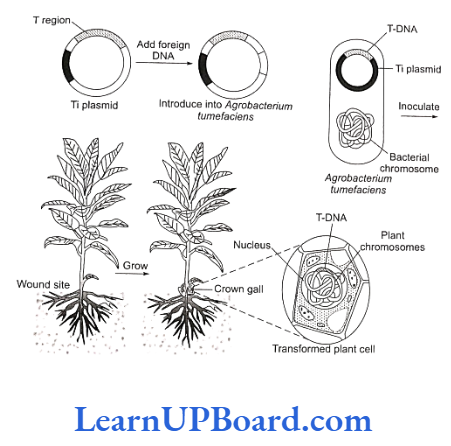
- Ti plasmid based gene transfer:
- A more common method of introducing foreign DNA into plant cells is to use the bacterium Agrobacterium tumefaciens and its Ti plasmid.
- This Gram-negative soil bacterium is a plant pathogen and produces crown gall disease in many dicotyledons including grapes, stone fruits, roses, tomato, sunflower, cotton, soybean, etc.
- Most strains of this bacterium carry Ti plasmid.
- In nature, Agrobacterium attaches to the leaves of plants and Ti plasmid is transferred into plant cells.
- The plasmid becomes incorporated into plant chromosomal DNA.
- Therefore, Ti plasmid has been used as a vehicle for the introduction of recombinant DNA into plant cells.
- Ti plasmids cause tumors in plants.
- Strains of the bacterium have been developed which do not have tumor-inducing genes. However, the T-region of plasmid plays an important role in gene transfer.
- This specific segment of bacterial plasmid DNA is called T-DNA (transferred DNA).
- T-DNA has a cloning site into which foreign DNA (DNA insert) is inserted.
- This recombinant plasmid is now introduced into bacterium Agrobacterium tumefaciens. It is then used to infect cultured cells.
- The T-region of the plasmid with foreign DNA (or DNA insert) is transferred to plant cells.
- It gets integrated with the chromosomal DNA of the cell.
- Cultured cells are induced to grow into plantlets.
- These are planted into the soil where mature plants are formed.
Identification of Recombinant
- After the insertion of recombinant DNA into the host cell, these need to be identified from those which do not possess it.
- The methods used to do so consider expression or non- expression of certain characters especially antibiotic resistance gene (e.g., ampicillin resistance gene) on plasmid vector.
- Selectable marker usually provides resistance against a substrate which when added to the culture medium inhibits the growth of normal cells or tissues in culture, so that only transformed tissues will grow.
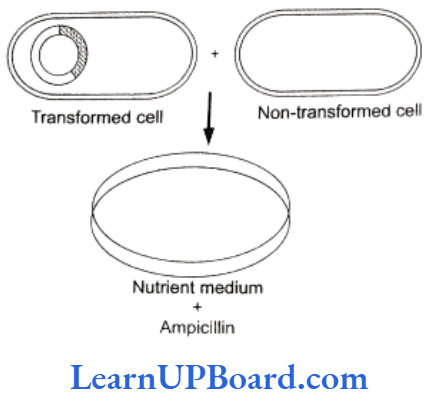
- Thus, the simplest method for identification is to grow transformed host cells (with ampicillin resistance gene) on medium containing ampicillin.
- This would enable the cells containing this trans- formed plasmid to grow and form colonies.
- There are other methods for the detection of recombinants based on the fact that the cloned DNA fragment disturbs the coding sequence of gene.
- This is known as insertional inactivation.
- Let us consider a plasmid containing genes resistant for two different antibiotics: ampicillin and tetracycline.
- If the target DNA fragment is inserted in a site located in ampicillin resistance gene, this gene will then be in-activated.
- Thus, host cells with such a recombinant plasmid will be sensitive to ampicillin but resistant to tetracycline.
- These host cells will die when grown on ampicillin containing medium but would grow on medium containing tetracycline.
- Self-ligated or religated (non-recombinant) vectors would grow on medium containing both ampicillin and tetracycline, being resistant to them.
- Another, but similar, method involves insertional inactivation of the lac Z gene.
- It is known as blue-white selection, being color based.
Gene Product Manufacture
- When recombinant DNA is transferred into a bacterial, plant, or animal cell, the foreign DNA is multiplied.
- Most of the recombinant technologies are aimed to produce a desirable protein.
- So, there is a need for expression recombinant DNA. After the cloning of the gene of interest, one has to maintain the optimum conditions to induce the expression of the target protein and consider producing it on a large scale.
- If any protein encoding gene is expressed in a heterologous host, it is known as a recombinant protein.
- The cells having cloned genes of interest can be grown on a small scale in the laboratory.
- Cultures may be used for extracting and purifying the desired protein.
- The cells can also be multiplied in a continuous system where the used medium is passed out from one side and the fresh medium is added from the other side to maintain the cells in their physiologically most active lag exponential phase (lag phase: no significant increase of cells; exponential phase: rapid multiplication of cells).
- This type of culturing method produces a larger biomass to get higher yields of the desired protein.
- Small volume cultures cannot give large quantities of products.
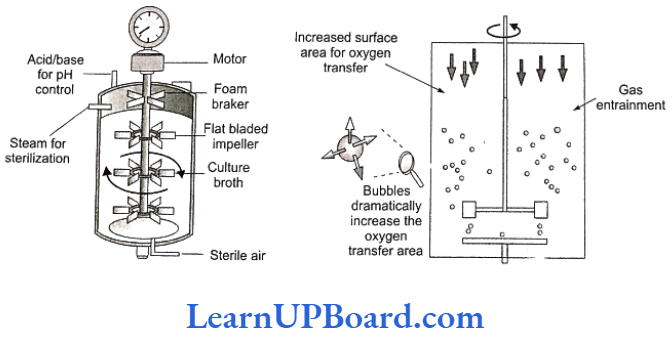
- To produce large quantities of these products, the development of bioreactors is required where large volumes (100-1000 L) of culture can be processed. Hence, bioreactors are like vessels in which raw materials are biologically converted into specific products (individual enzymes) using microbial, plant, animal, or human cells.
- A bioreactor provides the optimal conditions for obtaining the desired product by providing optimum growth conditions such as substrate, temperature, pH, vitamins, oxygen, and salts.
- One of the most commonly used bioreactor is of stirring type.
- A stirred reactor is usually cylindrical with a curved base to allow mixing of the contents of the reactor.
- The presence of stirrer makes mixing possible and also makes oxygen available through the reactor.
- A bioreactor also has an agitatory system, an oxygen delivery system, a foam control system, a temperature control system, pH control system, and sampling ports so that small volumes of culture can be withdrawn periodically.
Downstream Processing
- Once the product is ready, it has to be processed for commercial use.
- This requires purification and strict quality control to maintain the efficacy.
- The products based on biotechnology must ensure that they satisfy the consumer needs and are not harmful.
- Therefore, a thorough checking of products at each level of manufacture is done.
- The manufacturing process and the quality control methods vary with each product.
Choose the correct answer:
Question 1. 3′-5′ exonucleolytic degradation of DNA is performed by which enzyme?
- DNA polymerase
- Alkaline phosphatase
- DNA ligase
- RNA polymerase
Answer. 1. DNA polymerase
Question 2. The technique of gel electrophoresis was developed by
- Kary Mullis
- J.S. Chamberlain
- A. Tiselius
- F. Sanger
Answer. 3. A. Tiselius
Question 3. Which of the following dyes can be used to visualize nucleic acid after electrophoresis?
- Acridine orange
- Ethidium bromide
- Bromophenol blue
- Both (1) and (2)
Answer. 4. Both (1) and (2)
Question 4. Which of the following bonds are formed by the action of DNA ligase?
- Sugar-phosphate bond
- Phosphodiester bond
- Both (1) and (2)
- Phosphate-phosphate bond
Answer. 3. Both (1) and (2)
Question 5. Who were responsible for the isolation of “methylase” kind of enzyme from E. coli in 1960’s?
- Cohen and Boyer
- Banting and Best
- Linn and Arber
- Smith and Wilcox
Answer. 3. Linn and Arber
Question 6. The specific sequence recognized by “molecular scissors” is called
- Isomer
- Isobar
- Misnomer
- Palindrome
Answer. 4. Palindrome
Question 7. When a piece of DNA is digested with Eco RI, what kind of ends are created?
- Blunt ends
- Flush ends
- Cohesive ends
- Non-staggered ends
Answer. 3. Cohesive ends
Question 8. The sticky ends generated by the action of Eco RI on insert DNA facilitate the action of which enzyme?
- DNA polymerase
- Taq polymerase
- Alkaline phosphatase
- DNA ligase
Answer. 4. DNA ligase
Question 9. Which is incorrect with respect to DNA polymerase III?
- It requires ATP for polymerase action.
- It is required for PCR.
- It is more active than DNA polymerases I and II.
- It requires a pre-formed DNA template to work on.
Answer. 2. It is required for PCR.
Question 10. Which is not an application of modern biotechnology?
- Production of humulin
- Developing a DNA vaccine
- Gene therapy
- Production of cheese and butter
Answer. 4. Production of cheese and butter
Question 11. Which of the following cannot be related to biotechnology?
- Integration of natural science and organisms.
- Techniques to alter the chemistry of DNA.
- Introducing undesirable genes into the target organism.
- Maintenance of sterile ambience to enable the growth of only the desired microbes.
Answer. 3. Introducing undesirable genes into the target organism.
Question 12. Which of the following specific DNA sequence is responsible for initiating replication?
- Vector site
- Restriction enzymes action site
- Ori site
- Palindromic site
Answer. 3. Ori site
Question 13. Autonomously replicating, circular, extra chromosomal DNA of prokaryotic cell is called
- Satellite DNA
- Plasmid
- Recombinant DNA
- Nucleoid
Answer. 2. Plasmid
Question 14. Key tools to be involved in recombinant DNA technology are
A. Restriction enzymes
B. Polymerase enzyme
C. Ligase enzymes
D. Vectors
- (A) only
- (A) and (C) only
- (A), (B), and (C)
- (A), (B), (C), and (D)
Answer. 4. (A), (B), (C), and (D)
Question 15. The first restriction endonuclease to be discovered was
- Hind II
- Eco RI
- Bam HI
- Pst I
Answer. 1. Hind II
Question 16. Approximately, how many restriction enzymes have been isolated from the different (over 230) strains of bacteria?
- 300
- 600
- 750
- 900
Answer. 4. 900
Question 17. The conventional method for naming restriction enzymes is followed. In case of Eco RI, the “R” indicates
- Genus
- Species
- Name of the scientist
- Strain
Answer. 4. Strain
Question 18. The restriction endonuclease enzyme binds to the DNA and cuts
- Any one strand of the double helix
- Each of the two strands at specific points in their base-sugar bonds
- Each of the two strands at specific points in their base-phosphate bonds
- Each of the two strands at specific points in their sugar-phosphate backbones
Answer. 4. Each of the two strands at specific points in their sugar-phosphate backbones
Question 19. During gel electrophoresis, for the separation of DNA fragment, the
- Smallest fragment will move to the farthest point towards cathode
- Smallest fragment will move to the farthest point towards anode
- Largest fragment will move to the farthest point towards cathode
- Largest fragment will move to the farthest point towards anode
Answer. 2. Smallest fragment will move to the farthest point towards anode
Question 20. After electrophoresis, the separated DNA fragment can be visualized in ethidium bromide gel exposed to UV light. These DNA fragments appear as _________ colored bands.
- Orange
- Blue
- Silver
- Green
Answer. 1. Orange
Question 21. The procedure through which a piece of DNA is introduced in a host bacterium is called
- Cloning
- Transformation
- PCR
- Clonal selection
Answer. 2. Transformation
Question 22. After completing the transformation experiment involving the coding sequence of enzyme a-galactosidase, the recombinant colonies should
- Give blue color
- Not give blue color
- Have active α-galactosidase
- Both (2) and (3)
Answer. 2. Not give blue color
Question 23. Which of the following has the ability to transform normal cells into cancerous cells in animals?
- Agrobacterium tumifaciens
- Retroviruses
- DNA viruses
- Plasmids
Answer. 2. Retroviruses
Question 24. Which of the following is not applicable to Agrobacterium tumifaciens?
- Pathogen of several dicot plants.
- Has the ability to transform normal plant cells.
- Delivers gene of our interest.
- Ti plasmid of it is always pathogenic to plants with out any exception.
Answer. 4. Ti plasmid of it is always pathogenic to plants with out any exception.
Question 25. Insertional inactivation is related to
- Microinjection
- Gene gun
- Gel electrophoresis
- Selection of recombinants
Answer. 4. Selection of recombinants
Question 26. For transformation with recombinant DNA, the bacterial cells must first be made competent, which means
- Should increase their metabolic reactions
- Should decrease their metabolic reactions
- Increase efficiency with which DNA enters the bacterium
- Ability to divide fast
Answer. 3. Increase efficiency with which DNA enters the bacterium
Question 27. Which of the following method can be used for making the bacterial cell competent?
- Treating with specific concentration of divalent cation (Ca2+).
- Treating with specific concentration of monovalent cation (K).
- Heat shock.
- Both (1) and (3).
Answer. 4. Both (1) and (3).
Question 28. Which of the following techniques can be used to introduce foreign DNA into cell?
- Using disarmed pathogen
- Microinjection
- Gene gun
- All of these
Answer. 4. All of these
Question 29. During heat shock, the temperature used for giving thermal shock to the bacterium is
- 82°C
- 100°C
- Liquid nitrogen
- 42°C
Answer. 4. 42°C
Question 30. Which of the following enzymes is used in case of fungus to cause the release of DNA along with other macromolecules?
- Lysozyme
- Cellulase
- Chitinase
- Amylase
Answer. 3. Chitinase
Question 31. During the isolation of DNA, the addition of which of the following causes the precipitation of purified DNA?
- Chilled ethanol
- Ribonuclease enzyme
- DNA polymerase
- Proteases
Answer. 1. Chilled ethanol
Question 32. Which of the following is the correct sequence of PCR (polymerase chain reaction)?
- Denaturation → Annealing → Extension
- Extension → Denaturation → Annealing
- Annealing → Extension → Denaturation
- Denaturation → Extension → Annealing
Answer. 1. Denaturation → Annealing → Extension
Question 33. The most commonly used bioreactor is of stirring type. The stirrer facilitates
- Temperature control
- pH control
- Oxygen availability
- Product removal
Answer. 3. Oxygen availability
Question 34. After the completion of biosynthetic stage, the separation and purification of product is called
- Upstream processing
- Downstream processing
- Modern biotechnology
- Gene amplification
Answer. 2. Downstream processing
Question 35. From isolated DNA from a cell culture with seven de- sired genes, DNA segment can be excised by molecular scissors or chemical scalpels what biotechnologists call as
- Polymerase enzymes
- DNA ligase
- Restriction enzymes
- Helicase
Answer. 3. Restriction enzymes
Question 36. All the following statements about Stanley Cohen and Herbert Boyer are correct but one is wrong. Which one is wrong?
- They discovered recombinant DNA (rDNA) technology, and this marks the birth of modern biotechnology.
- They first produced healthy sheep clone, a Finn Dorset lamb, Dolly, from the differentiated adult mammary cells.
- They invented genetic engineering by combining a piece of foreign DNA containing a gene from a bacterium with a bacterial plasmid using the enzyme restriction endonuclease.
- They isolated the antibiotic resistance gene by cutting out a piece of DNA from the plasmid which was responsible for conferring antibiotic resistance.
Answer. 2. They first produced healthy sheep clone, a Finn Dorset lamb, Dolly, from the differentiated adult mammary cells.
Question 37. What is the fate of a piece of DNA which is somehow transferred into an alien organism?
- This piece of DNA would not be able to multiply itself in the progeny cells of the organism if not integrated into the genome of the organism.
- If the alien piece of DNA has become a part of the chromosome, it will replicate.
- If the alien piece of DNA is linked with the origin of replication in chromosome, it will replicate.
- All of these.
Answer. 4. All of these.
Question 38. In the year 1963, the two enzymes responsible for restricting the growth of bacteriophage in Escherichia coli were isolated. These were and, respectively.
- Ligase, restriction endonuclease
- Helicase, restriction endonuclease
- Methylase, restriction endonuclease
- DNA polymerase, restriction endonuclease
Answer. 3. Methylase, restriction endonuclease
Question 39. The cutting of DNA by restriction endonucleases results in fragments of DNA. These fragments are generally separated by a technique known as
- Gel-filtration chromatography
- Centrifugation
- Gel electrophoresis
- Thin-layer chromatography
Answer. 3. Gel electrophoresis
Question 40. Which of the following bacteria are known as natural genetic engineers of plants, as gene transfer is happening in nature without human interference?
- Azotobacter
- Agrobacterium tumefaciens
- Escherichia coli
- Rhizobium
Answer. 2. Agrobacterium tumefaciens
Question 41. The technique in which a foreign DNA is precipitated on the surface of tungsten or gold particles and shot into the target cells is known as
- Microinjection
- Chemical-mediated genetic transformation
- Electroporation
- Biolistics
Answer. 4. Biolistics
Question 42. The isolation of the genetic material in pure form-free from other macromolecules can be achieved by treating the bacterial cells/plant or animal tissues with the following enzymes, except
- Lysozyme
- Cellulase
- Chitinase
- Ligase
Answer. 4. Ligase
Question 43. Which of the following is not a recombinant protein used in medical practice?
- TPA (tissue plasminogen activator)
- Interferon (α, B, and 7)
- Vaccine (for hepatitis B)
- Heparin
Answer. 4. Heparin
Question 44. cDNA is
- Circular DNA in bacteria
- Complementary DNA
- Copy DNA
- Both (2) and (3)
Answer. 4. Both (2) and (3)
Question 45. The Noble Prize of 1978 for restriction endonuclease technology was given to
- Temin and Baltimore
- Milstein and Kohler
- Arber, Nathans, and Smith
- Holley, Khorana, and Nirenberg
Answer. 3. Arber, Nathans, and Smith
Question 46. Plasmids are used in genetic engineering because they
- Are easily available
- Are able to integrate with host chromosome
- Are able to replicate along with chromosomal DNA
- Contain DNA sequence coding for drug resistance
Answer. 3. Are able to replicate along with chromosomal DNA
Question 47. Which of the following processes and techniques are included under biotechnology?
A. In vitro fertilization leading to a test-tube baby.
B. Synthesizing gene and using it.
C. Developing DNA vaccine.
D. Correcting a defective gene.
- (B) and (D) only (3)
- (B), (C), and (D)
- (A) and (B)
- (A), (B), (C), and (D)
Answer. 4. (A), (B), (C), and (D)
Question 48. The tumor-inducing (Ti) plasmid has now been modified to a cloning vector which is no more pathogenic to plants but is still able to use mechanisms to deliver genes of our interest into a variety of plants because Ti plasmid has been modified by
- Adding tumor-forming genes
- Deleting tumor-forming genes
- Adding genes resistant to endonucleases
- Deleting endonuclease
Answer. 2. Deleting tumor-forming genes
Question 49. Which of the following statements is incorrect?
- Plasmids have the ability to replicate within the bacterial cells independent of the control of chromosomal DNA.
- Some plasmids have only one or two copies per cell whereas others may have 15-100 copies per cell.
- Bacteriophages have the ability to replicate within the bacterial cell independent of the control of chromosomal DNA.
- Transformation is a procedure of separation and isolation of DNA fragments.
Answer. 4. Transformation is a procedure of separation and isolation of DNA fragments.
Question 50. Which of the following is the first artificial cloning vector that has two selectable markers-tetracycline (tetR) and antibiotic restriction enzymes (ampR)?
- YAC
- BAC
- pBR322
- Cosmid vectors
Answer. 3. pBR322
Question 51. Restriction endonucleases are most widely used in recombinant DNA technology. They are obtained from
- Bacteriophage
- Bacterial cells
- Plasmids
- All prokaryotic cells
Answer. 2. Bacterial cells
Question 52. All the following statements are correct about genetic engineering, but one is wrong. Which one is wrong?
- It is a technique for artificially and deliberately modifying DNA (genes) to suit human needs.
- It is often referred as gene splicing.
- The organism carrying the foreign genes is termed as transgenic or GMO.
- Alec Jeffrey is the father of genetic engineering.
Answer. 4. Alec Jeffrey is the father of genetic engineering.
Question 53. All the following are the properties of enzyme Taq polymerase, except
- It is thermostable DNA polymerase
- It is isolated from a bacterium, Thermus aquaticus
- It is used for the amplification of gene of interest using PCR
- It is thermostable RNA polymerase
Answer. 4. It is thermostable RNA polymerase
Question 54. Which of the following is incorrect match?
- Gene therapy: An abnormal gene is replaced by normal gene
- Cloning: Ability to multiply copies of antibiotic resistance gene in E. coli
- Restriction enzymes: Molecular scissors
- Exonucleases: Molecular glue
Answer. 4. Exonucleases: Molecular glue
Question 55. Appropriate techniques have been developed for large- scale cell culture using bioreactors for producing
- Foreign gene product
- Vaccines
- Hormones
- All of these
Answer. 4. All of these
Question 56. The uptake of genes by cells in microbes and plants is termed as
- Insertional inactivation
- Transformation
- Selectable markers
- Cloning vectors
Answer. 2. Transformation
Question 57. If we ligate a foreign DNA at the BamHI site of tetracycline resistance gene in pBR322, the recombinant plasmid will
- Show ampicillin resistance only
- Show tetracycline resistance
- Will grow well on tetracycline-containing medium
- Will not grow on ampicillin-containing medium
Answer. 1. Show ampicillin resistance only
Question 58. Polyethylene glycol can help in the uptake of foreign DNA into the host cell. This type of gene transfer is called
- Electroporation
- Chemical mediated genetic transformation
- Microinjection
- Particle gun
Answer. 2. Chemical mediated genetic transformation
Question 59. The normal E. coli cells carry resistance against which of the following antibiotics?
- Ampicillin
- Chloramphenicol
- Tetracycline or kanamycin
- None of these
Answer. 4. None of these
Question 60. The isolation of genetic material from fungal cells does not involve the use of
- Agarose
- Chitinase
- Ethanol
- Water
Answer. 1. Agarose
Question 61. In a restriction digestion experiment, the sticky ends of vector rejoined forming a circular vector without insert. Which enzyme can be used to eliminate this possibility?
- DNA ligase
- Alkaline phosphatase
- DNA polymerase
- RNA polymerase
Answer. 2. Alkaline phosphatase
Question 62. Denaturation can be achieved at which temperature during PCR?
- 72°C
- 95°C
- 40°C
- 25°C
Answer. 2. 95°C
Question 63. Choose the incorrect statement with respect to Agrobacterium tumefaciens.
- It is a Gram-negative soil bacterium.
- It produces crown gall disease in dicot plants.
- The foreign DNA is inserted at the ori site of Ti plasmid.
- Ti plasmid becomes incorporated into the plant chromosomal DNA.
Answer. 3. The foreign DNA is inserted at the ori site of Ti plasmid.
Question 64. Which is not a method for the introduction of recombinant DNA into host cells?
- Electroporation
- Biolistics
- Transfection
- Restriction digestion
Answer. 4. Restriction digestion
Question 65. The essential requirements for a gene amplification reaction are
- 20 mg of DNA template
- Forward and reverse primers
- Mg2+
- All of these
Answer. 4. All of these
Question 66. Choose the incorrect statement with respect to PCR reaction:
- It requires Taq polymerase.
- It requires dNTP’s.
- It generates 2n molecules after n number of cycles.
- The optimum temperature for polymerization step is greater than or equal to 90°C.
Answer. 4. The optimum temperature for polymerization step is greater than or equal to 90°C.
Question 67. Which is not an application of PCR?
- DNA fingerprinting
- DNA foot-printing
- Detection of mutation
- Prenatal diagnosis
Answer. 2. DNA foot-printing
Question 68. Rejoining of vector molecule after restriction enzyme digestion can be avoided by
- Using different enzymes for insert and vector
- Using same enzyme for insert and vector
- Using DNA ligase immediately after digestion
- Using alkaline phosphatase on only vector
Answer. 4. Using alkaline phosphatase on only vector
Question 69. If the target gene is inserted at Sal 1 site of the recombinant, plasmid will show resistance for pBR322
- Ampicillin
- Tetracyline
- Both (1) and (2)
- Kanamycin
Answer. 1. Ampicillin
Question 70. It is theoretically possible for a gene from any organism to function in any other organism. Why is this possible?
- All organisms have ribosomes.
- All organisms have the same genetic code.
- All organisms are made up of cells.
- All organisms have similar nuclei.
Answer. 2. All organisms have the same genetic code.
Question 71. If you discovered a bacterial cell that contained no restriction enzymes, which of the following would you except to happen?
- The cell would create incomplete plasmids.
- The cell would be unable to replicate its DNA.
- The cell would become an obligate parasite.
- The cell would be easily infected by bacteriophages.
Answer. 4. The cell would be easily infected by bacteriophages.
Question 72. Assume that you are trying to insert a gene into a plasmid and someone gives you a preparation of DNA cut with restriction enzyme X. The gene you wish to insert has sites on both ends for cutting by restriction enzyme Y. You have a plasmid with a single site for Y, but not for X. Your strategy should be to
- Cut the plasmid with restriction enzyme X and insert the fragments cut with Y into the plasmid
- Cut the plasmid with restriction enzyme X and insert the gene into the plasmid
- Cut the plasmid twice with restriction enzyme Y and ligate the two fragments into the plasmid cut with the same enzyme
- Cut the plasmid twice with restriction enzyme Y and ligate the two fragments onto the ends of the human DNA fragments cut with restriction enzyme X
Answer. 3. Cut the plasmid twice with restriction enzyme Y and ligate the two fragments into the plasmid cut with the same enzyme
Question 73. 1. Transform bacteria with recombinant DNA molecule.
2. Cut the plasmid DNA using restriction enzymes.
3. Extract plasmid DNA from bacterial cells.
4. Hydrogen-bond the plasmid DNA to non-plasmid DNA fragments.
5. Use ligase to seal plasmid DNA to non-plasmid DNA.
From the given list, which of the following is the most logical sequence of steps for splicing foreign DNA into a plasmid and inserting the plasmid into a bacterium?
- 4, 5, 1, 2, 3
- 3, 2, 4, 5, 1
- 3, 4, 5, 1, 2
- 2, 3, 5, 4, 1
Answer. 2. 3, 2, 4, 5, 1
Question 74. A eukaryotic gene has sticky ends produced by restriction endonuclease Eco RI. The gene is added to a mixture containing Eco RI and a bacterial plasmid that carries two genes, which make it resistant to ampicillin and tetracyline. The plasmid has one recognition site for Eco RI located in the tetracycline resistance gene. This mixture is incubated for several hours and then added to bacteria growing in nutrient broth. The bacteria are allowed to grow overnight and are streaked on a plate using a technique which produces isolated colonies that are clones of the original. Samples of these colonies are then grown in four different media: nutrient broth plus ampicillin, nutrient broth plus tetracycline, nutrient broth plus ampicillin and tetracycline, and nutrient broth containing no antibiotics.
The bacteria containing the engineered plasmid would grow in
- The ampicillin and tetracycline broth only
- The nutrient broth, the ampicillin broth, and the tetracycline broth
- The nutrient broth and the ampicillin broth only
- The nutrient broth only
Answer. 3. The nutrient broth and the ampicillin broth only
Question 75. Agrobacterium tumefaciens is used in genetic engineering for
- DNA mapping
- DNA modification
- Vector
- DNA fingerprinting
Answer. 3. Vector
Question 76. A genetically engineered bacteria used for clearing oil spills is
- Escherischia coli
- Bacillus subtilis
- Agrobacterium tumefaciens
- Pseudomonas putida
Answer. 4. Pseudomonas putida
Question 77. Who isolated the first restriction endonucleases?
- Temin and Baltimore
- Sanger
- Nathans and Smith
- Paul Berg
Answer. 3. Nathans and Smith
Question 78. Genetic engineering is
- Study of extra-nuclear gene
- Manipulation of genes by artificial method
- Manipulation of RNA
- Manipulation of enzymes
Answer. 2. Manipulation of genes by artificial method
Question 79. Which of the following enzymes cut the DNA molecule at specific nucleotide sequence?
- Restriction endonuclease
- DNA ligase
- RNA polymerase
- Exonuclease
Answer. 1. Restriction endonuclease
Question 80. DNA fingerprinting was invented by
- Karl Mullis
- Alec Jeffery
- Dr. Paul Berg
- Francis Collins
Answer. 2. Alec Jeffery
Question 81. Which structure is involved in genetic engineering?
- Plastid
- Plasmid
- Codon
- None
Answer. 2. Plasmid
Question 82. Which of the following is the example of chemical scissors?
- Eco RI
- HindIII
- Bam I
- All of the above
Answer. 4. All of the above
Question 83. Restriction endonucleases are used in genetic engineering because
- They can degrade harmful proteins
- They can join DNA fragments
- They can cut DNA at variable sites
- They can cut DNA at specific base sequences
Answer. 4. They can cut DNA at specific base sequences
Question 84. Chimeric DNA is
- DNA which contains uracil
- DNA synthesized from RNA
- Recombinant DNA
- DNA which contains single strand
Answer. 3. Recombinant DNA
Question 85. A piece of nucleic acid used to find out a gene by forming hybrid with it is called
- cDNA
- DNA probe
- Sticky end
- Blunt end
Answer. 2. DNA probe
Question 86. Which of the following is the example of direct gene transfer?
- Microinjection
- Electroporation
- Particle gun
- All the above
Answer. 4. All the above
Question 87. How many copies of DNA sample are produced in PCR technique after 6 cycles?
- 4
- 32
- 16
- 64
Answer. 3. 16
Question 88. The basic procedure involved in the synthesis of recombinant DNA molecule is depicted. The mistake in the cedure is procedure is
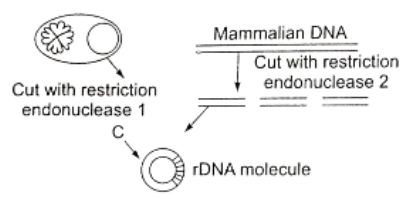
- Enzyme polymerase is not included
- The mammalian DNA is shown double stranded
- Only one fragment is inserted
- Two different restriction enzymes are used
Answer. 4. Two different restriction enzymes are used
Question 89. Western blotting is used for the identification of
- DNA
- RNA
- Protein
- All the above
Answer. 3. Protein
Question 90. In rDNA technique, which of the following technique is not used in introducing DNA into host cell?
- Transduction
- Conjugation
- Transformation
- Electroporation
Answer. 2. Conjugation
Question 91. Which of the following techniques are used in analyzing restriction fragment length polymorphism (RFLP)?
(a) Electrophoresis
(b) Electroporation
(c) Methylation
(d) Restriction digestion
- (a) and (c)
- (c) and (d)
- (a) and (d)
- (b) and (d)
Answer. 3. (a) and (d)
Question 92. Restriction enzymes are
- Not always required in genetic engineering
- Essential tool in genetic engineering
- Nucleases that cleave DNA at specific sites
- (2) and (3) both
Answer. 4. (2) and (3) both
Question 93. The function of restriction endonuclease enzyme is
- It is useful in genetic engineering
- It protects the bacterial DNA against foreign DNA
- It is helpful in transcription
- It is helpful in protein synthesis
Answer. 2. It protects the bacterial DNA against foreign DNA
Question 94. A bacterium modifies its DNA by adding methyl groups to the DNA. It does so to
- Clone its DNA
- Be able to transcribe many genes simultaneously
- Turn its gene on
- Protect its DNA from its own restriction enzyme
Answer. 4. Protect its DNA from its own restriction enzyme
Question 95. Plasmid has been used as vector because
- It is circular DNA which has capacity to join eukaryotic DNA
- It can move between prokaryotic and eukaryotic cells
- Both ends show replication
- It has antibiotic resistance gene
Answer. 1. It is circular DNA which has capacity to join eukaryotic DNA
Question 96. Which of the following cuts the DNA from specific places?
- Restriction endonuclease (Eco RI)
- Ligase
- Exonuclease
- Alkaline phosphate
Answer. 1. Restriction endonuclease (Eco RI)
Question 97. The manipulation of DNA in genetic engineering becomes possible due to the discovery of
- Restriction endonuclease
- DNA ligase
- Transcriptase
- Primase
Answer. 1. Restriction endonuclease
Question 98. Restriction enzymes
- Are endonucleases which cleave DNA at specific sites
- Make DNA complementary to an existing DNA or RNA
- Cut or join DNA fragments
- Are required in vector-less direct gene transfer
Answer. 1. Are endonucleases which cleave DNA at specific sites
Question 99. DNA fingerprinting refers to
- Techniques used for the identification of fingerprints of individuals
- Molecular analysis of profiles of DNA samples
- Analysis of DNA samples using imprinting devices
- Techniques used for molecular analysis of different specimens of DNA
Answer. 2. Molecular analysis of profiles of DNA samples
Question 100. Restriction endonucleases
- Are synthesized by bacteria as part of their defense compound
- Are present in mammalian cells for degradation of DNA when the cell dies
- Are used in genetic engineering for ligating two DNA molecules
- Are used for in vitro DNA synthesis
Answer. 1. Are synthesized by bacteria as part of their defense compound
Question 101. What is the first step in the Southern blot technique?
- Denaturation of DNA on the gel for hybridization with specific probe.
- Production of a group of genetically identical cells.
- Digestion of DNA by restriction enzyme.
- Isolation of DNA from a nucleated cell such as the one from the scene of crime.
Answer. 1. Denaturation of DNA on the gel for hybridization with specific probe.
Question 102. Which of the following is not produced by E. coli in lactose?
- B-galactosidase
- Thiogalactoside transacetylase
- Lactose dehydrogenase
- Lactose permease
Answer. 3. Lactose dehydrogenase
Question 103. The technique of transferring DNA fragment separated on agarose gel to a synthetic membrane such as nitrocellulose is known as
- Northern blotting
- Southern blotting
- Western blotting
- Dot blotting
Answer. 2. Southern blotting
Question 104. The production of a human protein in bacteria in genetic engineering is possible because
- Bacterial cell can carry out RNA splicing reactions
- The mechanism of gene regulation is identical in humans and bacteria
- Human chromosome can replicate in bacterial cell
- Genetic code is universal
Answer. 4. Genetic code is universal
Question 105. Electroporation procedure involves
- Fast passage of food through sieve pores in phloem elements with the help of electric stimulation
- Opening of stomatal pores during night by artificial light
- Making transient pores in the cell membrane to introduce gene constructs
- Purification of saline water with the help of a membrane system
Answer. 3. Making transient pores in the cell membrane to introduce gene constructs
Question 106. The total number of nitrogenous bases in human genome is estimated to be about
- 3.5 million
- 35 thousand
- 35 million
- 3.1 billion
Answer. 4. 3.1 billion
Question 107. Two microbes found to be very useful in genetic engineering are
- Escherichia coli and Agrobacterium tumefaciens
- Vibro cholerae and a tailed bacteriophage
- Diplococcus sp. and Pseudomonas sp.
- Crown gall bacterium and Caenorhabdits elegans
Answer. 1. Escherichia coli and Agrobacterium tumefaciens
Question 108. Restriction endonuclease
- Cuts the DNA molecule randomly
- Cuts the DNA molecule at specific sites
- Restricts the synthesis of DNA inside the nucleus
- Synthesizes DNA
Answer. 2. Cuts the DNA molecule at specific sites
Question 109. The restriction enzyme Eco RI has the property of
- Endonuclease activity
- Exonuclease activity
- Ligation activity
- Correcting the topology of replicating DNA
Answer. 1. Endonuclease activity
Question 110. DNA ligase is an enzyme that catalyzes the
- Splitting of DNA threads into small bits
- Joining of the fragments of DNA
- Denaturation of DNA
- Synthesis of DNA
Answer. 2. Joining of the fragments of DNA
Question 111. More advancement in genetic engineering is due to
- Restriction endonuclease
- Reverse transcriptase
- Protease
- Zymase
Answer. 1. Restriction endonuclease
Question 112. The function of PCR is
- Translation
- Transcription
- DNA amplification
- None of these
Answer. 3. DNA amplification
Question 113. Which of the following is used as the best genetic vector?
- Bacillus thuriengenesis
- Agrobacterium tumefaciens
- Pseudomonas putida
- All of these
Answer. 2. Agrobacterium tumefaciens
Question 114. The transfer of protein from electrophoretic gel to nitrocellulose membrane is known as
- Transferase
- Northern blotting
- Western blotting
- Southern blotting
Answer. 3. Western blotting
Question 115. DNA fingerprinting was first discovered by
- Alec Jeffery
- Cark Mullis
- C. Milstein
- Dr. Paul Berg
Answer. 1. Alec Jeffery
Question 116. Which of the following enzyme is used to join DNA fragments?
- Terminase
- Endonuclease
- Ligase
- DNA polymerase
Answer. 3. Ligase
Question 117. Restriction endonucleases are enzymes which
- Make cuts at specific positions within the DNA molecule
- Recognize a specific nucleotide sequence for binding of DNA ligase
- Restrict the action of enzyme DNA polymerase
- Remove nucleotides from the ends of the DNA molecule
Answer. 1. Make cuts at specific positions within the DNA molecule
Question 118. Satellite DNA is a useful tool in
- Organ transplantation
- Sex determination
- Forensic science
- Genetic engineering
Answer. 3. Forensic science
Question 119. PCR and restriction fragment length polymorphism are the methods for
- Genetic transformation
- DNA sequencing
- Genetic fingerprinting
- Study of enzymes
Answer. 3. Genetic fingerprinting
Question 120. Which one is a true statement regarding DNA polymerase used in PCR?
- It serves as a selectable marker.
- It is isolated from a virus.
- It remains active at high temperature.
- It is used to ligate introduced DNA in recipient cells.
Answer. 3. It remains active at high temperature.
Question 121. For transformation, microparticles coated with DNA to be bombarded with gene gun are made up of
- Platinum or zinc
- Silicon or platinum
- Gold or tungsten
- Silver or platinum
Answer. 3. Gold or tungsten
Question 122. A single strand of nucleic acid tagged with a radioactive molecule is called
- Selectable marker
- Plasmid
- Probe
- Vector
Answer. 3. Probe
Question 123. The following figure is the diagrammatic representation of the E. coli vector PBR 322. Which one of the given options correctly identifies its certain component(s)?
- rop-reduced osmotic pressure
- Hind III, Eco RI-selectable markers
- ampR, tetR-antibiotic resistance genes
- ori-original restriction enzyme
Answer. 3. ampR, tetR-antibiotic resistance genes
Question 124. Which one of the following is a case of wrong matching?
- Vector DNA: Site for tRNA synthesis
- Micropropagation: In vitro production of plants in large numbers
- Callus: Unorganized mass of cells produced in tissue culture
- Somatic hybridization: Fusion of two diverse cells
Answer. 1. Vector DNA: Site for tRNA synthesis
Question 125. Biolistics (gene gun) is suitable for
- Disarming pathogen vectors
- Transformation of plant cells
- Constructing recombinant DNA by joining with vectors
- DNA fingerprinting
Answer. 3. Constructing recombinant DNA by joining with vectors
Question 126. In genetic engineering, the antibiotics are used
- As selectable markers
- To select healthy vectors
- As sequences from where replication starts
- To keep the cultures free of infection
Answer. 1. As selectable markers
Question 127. What is it that forms the basis of DNA fingerprinting?
- The relative proportions of purines and pyrimidines in DNA.
- The relative difference in the DNA occurrence in blood, skin, and saliva.
- The relative amount of DNA in the ridges and grooves of fingerprints.
- Satellite DNA occurring as highly repeated short DNA segments.
Answer. 4. Satellite DNA occurring as highly repeated short DNA segments.
Question 128. The following figure shows three steps (A)-(C) of PCR. Select the option giving correct identification together with what it represents?

Options:
- B-Denaturation at a temperature of about 98°C separating the two DNA strands
- A-Denaturation at a temperature of about 50°C
- C-Extension in the presence of heat stable DNA polymerase
- A-Annealing with two sets of primers
Answer. 3. C-Extension in the presence of heat stable DNA polymerase
Question 129. Which one of the following represents a palindromic sequence in DNA?
- 5′-GAATTC-3′; 3′- CTTAAG-5′
- 5′-CCATCC-3′; 3′-GAATCC-5′
- 5′-CATTAG-3′; 3′-GATAAC-5′
- 5′-GATACC-3′; 3′-CCTAAG-5′
Answer. 1. 5′-GAATTC-3′; 3′- CTTAAG-5′
Question 130. DNA fragments generated by restriction endonucleases in a chemical reaction can be separated by
- Polymerase chain reaction
- Electrophoresis
- Restriction mapping
- Centrifugation
Answer. 2. Electrophoresis
Question 131. The colonies of recombinant bacteria appear white in contrast to the blue colonies of non-recombinant bacteria because of
- Insertional inactivation of alpha-galactosidase in non-recombinant bacteria
- Insertional inactivation of alpha-galactosidase in recombinant bacteria
- Inactivation of glycosidase enzyme in recombinant bacteria
- Non-recombinant bacteria containing beta-galactosidase
Answer. 4. Non-recombinant bacteria containing beta-galactosidase
Assertion-Reasoning Questions
In the following questions, a statement of Assertion (A) is followed by a statement of Reason (R).
- If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark (1).
- If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
- If Assertion is true but Reason is false, then mark (3).
- If both Assertion and Reason are false, then mark (4).
Question 1. Assertion: DNA ligase plays important role in recombinant DNA technology.
Reason: The linking of antibiotic resistance gene with plasmid vector became possible by enzyme DNA ligase.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark(1).
Question 2. Assertion: Restriction enzymes belong to a larger class of enzymes called nucleases.
Reason: Each restriction enzyme recognizes a specific palindromic nucleotide sequence in the DNA.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 3. Assertion: During gel electrophoresis, DNA fragments move towards the anode.
Reason: DNA fragments are negatively charged molecules.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark(1).
Question 4. Assertion: The selection of recombinants due to inacti- vation of antibiotics is a cumbersome procedure.
Reason: It requires simultaneous plating on two plates having different antibiotics.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark(1).
Question 5. Assertion: Taq polymerase is involved in PCR technique.
Reason: This enzyme remains active during high temperature including denaturation of double-stranded DNA.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark(1).
Question 6. Assertion: Small DNA fragments will arrange towards the positive end after gel electrophoresis in DNA test.
Reason: DNA is negatively charged.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark(1).
Question 7. Assertion: PCR technique is used in the amplification of a specific gene.
Reason: In PCR technique, Taq polymerase enzyme is used, and this enzyme is thermosensitive.
Answer. 3. If Assertion is true but Reason is false, then mark (3).
