NEET Biology Notes Molecular Basis Of Inheritance
DNA
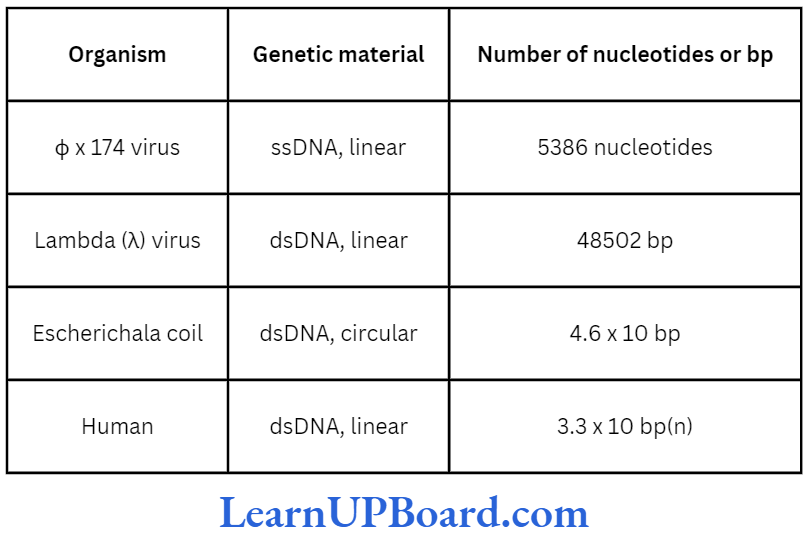
- DNA (deoxyribose nucleic acid) is a long polymer of deoxyribonucleotides.
- The length of DNA is usually defined as the number of nucleotides (or a pair of nucleotide referred to as base pairs or bp) present in it.
- This is the characteristic of an organism.
- A few examples are sited in table.
Chemical Structure of DNA Polynucleotide
- The basic unit of DNA is a nucleotide which has three components: a nitrogenous base, a pentose sugar (deoxyribose), and a phosphate group.
- There are two types of nitrogenous bases: Purines (adenine and guanine) and pyrimidines (cytosine and thymine).
- Cytosine is common for both DNA and RNA (ribose nucleic acid), and thymine is present in DNA only.
- Uracil is present in RNA at the place of thymine.
- A nitrogenous base is linked to the pentose sugar through an N-glycosidic linkage to form a nucleoside such as adenosine or deoxyadenosine, guanosine or deoxyguanosine, cytidine or deoxycytidine, and deoxythymidine.
- When a phosphate group is linked to 5′-OH of a nucleoside through phosphoester linkage, a corresponding nucleotide (or deoxynucleotide depending on the type of sugar present) is formed.
- Two nucleotides are linked through 3′-5′ phosphodiester linkages to form a dinucleotide.
- More nucleotides can be joined in such a manner to form a polynucleotide chain.
- The polymer, thus, formed has a free phosphate moiety at the 5′-end of sugar, which is referred to as the 5′-end of polynucleotide chain.
Read and Learn More NEET Biology Notes
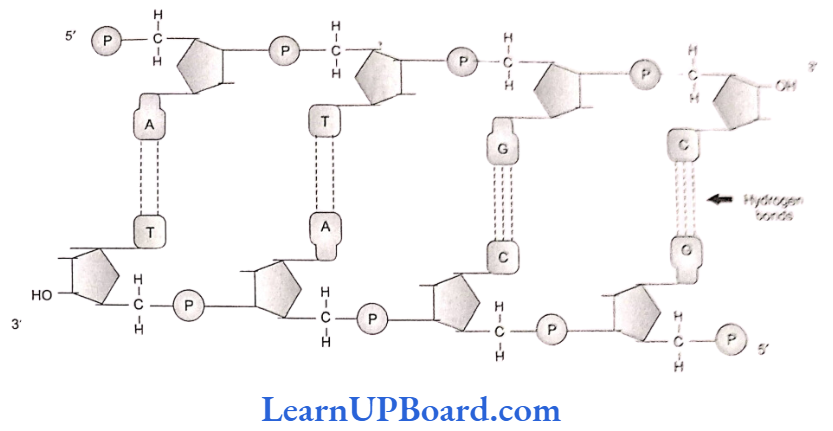
- Similarly, at the other end of the polymer, the sugar has a free 3′-OH group which is referred to as the 3′-end of polynucleotide chain.
- Backbone in a polynucleotide chain is formed due to sugar and phosphates (phosphodiester bond). Nitrogenous bases linked to the sugar moiety project from the backbone.
- In RNA, every nucleotide residue has an additional -OH group present at the 2′-position in the ribose.
- Also, in RNA, uracil is found at the place of thymine (S-methyluracil, another chemical name for thymine). In 1953, James Watson and Francis Crick, based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed double helix model for the structure of DNA.
- One of the hallmarks of their proposition was “base pairing between the two strands of polynucleotide chain.” However, this proposition was also based on the observation of Erwin Chargaff (that for a double-stranded DNA, the ratios between adenine and thymine and that between guanine and cytosine are constant and equal.
- Chargaff (1950) made observations on the bases and other contents of DNA. These observations or generalizations are known as Chargaff’s rules.
- Purine and pyrimidine base pairs are present in equal amounts, i.e., adenine (A) + guanine (G) = thymine (T) + cytosine (C).
- The molar amount of purine (adenine) is always equal to the molar amount of pyrimidine (thy- mine). Similarly, guanine is equalled by cytosine.
- Deoxyribose (sugar) and phosphate occur in equimolar proportions.
- The ratio (A+T)/(G+C) is constant for a species. It is called base ratio. It is 1.52 for humans and 0.93 for E. coli.
- Base pairing is a very unique property of polynucleotide chains.
- Both strands of DNA are said to be complementary to each other. Therefore, if the sequence of bases in one strand is known, then the sequence in the other strand can be predicted.
- Thus, if one DNA strand has A, the other will have T; and if one has G, the other will have C.
- Therefore, if the base sequence of one strand is CAT TAG GAC, the base sequence of the other strand will be GTA ATC CTG.
- Hence, the two polynucleotide strands are complementary to one another.
- Also, if each strand from a DNA or parental DNA acts as a template for the synthesis of a new strand, the two double-stranded DNA or daughter DNA produced will be identical to the parental DNA molecule.
Salient Features of DNA Double Helix
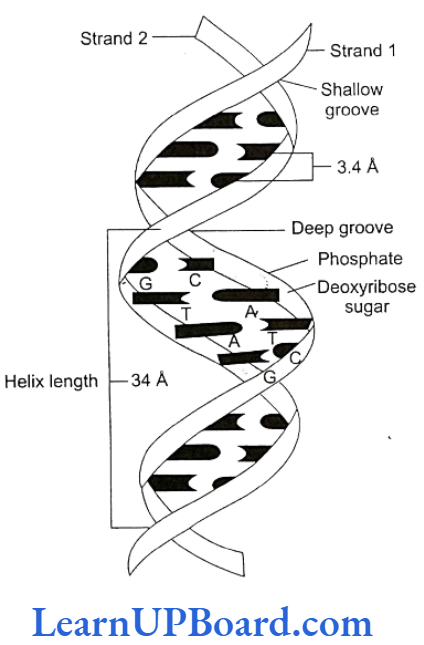
- DNA double helix is made of two polynucleotide chains, where the backbone is constituted by sugar- phosphate and the bases project inside.
- The two chains have anti-parallel polarity. It means, if one chain has polarity 5’P→ 3’OH, the other has 3’OH→ 5’P.
- The bases in the two strands are paired through hydrogen bonds (H-bonds), forming base pairs. Ad- enine forms two hydrogen bonds with thymine from the opposite strand and vice versa. Guanine is bonded with cytosine with three H-bonds. As a result, always a purine comes opposite to a pyrimidine. This generates approximately uniform distance between the two strands of the helix.
- The plane of one base pair stacks over the other in double helix. This, in addition to H-bonds, confers stability to the helical structure.
- The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm and there are roughly 10 bp in each turn. Consequently, the distance between the base pairs in a helix is approximately equal to 0.34 nm. It is because of specific base pairing with a purine lying opposite to pyrimidine which makes two chains 2 nm thick.
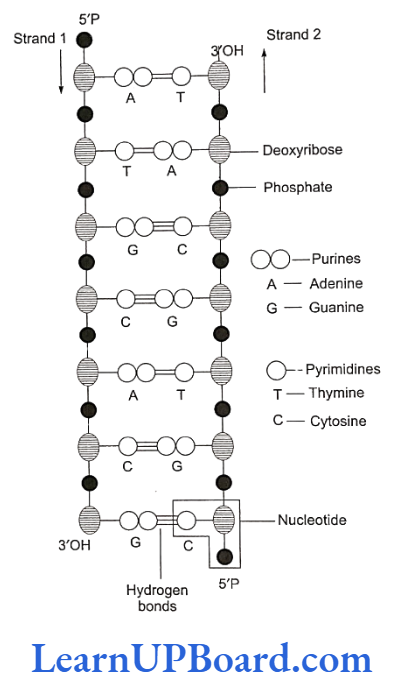
molecular basis of inheritance
NEET Biology Notes Molecular Basis Of Inheritance Packaging Of DNA Helix
- The average distance between two adjacent base pairs is 0.34 nm (0.34 x 10m or 3.4 A).
- The length of DNA for a human diploid cell is 6.6 x 10° bp x 0.34 x 10 m/bp = 2.2 m.
- This length is far greater than the dimension of a typical nucleus (approximately 10 m).
- The number of base pairs in E. coli is 4.6 × 10°. Total length is 1.36 mm.
- The long-sized DNA is accommodated in a small area (about 1 um in E. coli) only through packing or com- paction.
- DNA is acidic due to the presence of a large number of phosphate groups.
- Compaction occurs by folding and attachment of DNA with basic proteins-polyamine in prokaryotes and histone in eukaryotes.
DNA Packaging in Prokaryotes
- DNA is found in cytoplasm in supercoiled state.
- The coils are maintained by non-histone basic proteins such as polyamines.
- RNA may also be involved. This compact structure of DNA is called nucleoid or genophore.
DNA Packaging in Eukaryotes
- DNA packaging in eukaryotes is carried out with the help of lysine- and arginine-rich basic proteins called histones.
- The unit of compaction is called nucleosome.
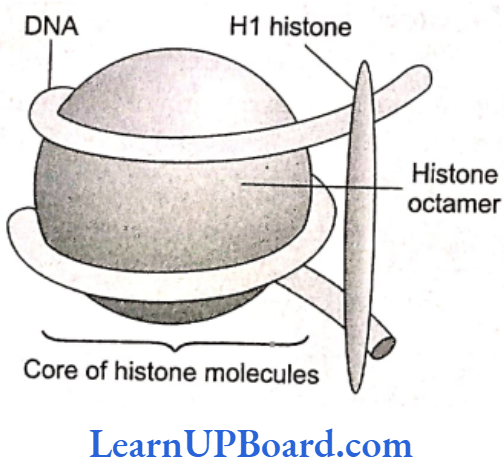
- There are five types of histone proteins: H1, H2A, H2B, H3, and H4.
- Four of these occur in pairs to produce histone octamer (two copies of each-H2A, H2B, H3, and H1) called nu-body or core of nucleosome.
- Their positively charged ends are directed outside.
- They attract negatively charged strands of DNA.
- About 200 bp of DNA are wrapped over nu-body to complete about 13 turns.
- This forms a nucleosome of size 110 x 60 Å (11 x 6 nm).
- The DNA present between two adjacent nucleosomes is called linker DNA.
- It is attached to H, histone protein.
- The length of linker DNA varies from species to species.
- Nucleosome chain gives a “beads-on-string” appearance under electron microscope ).
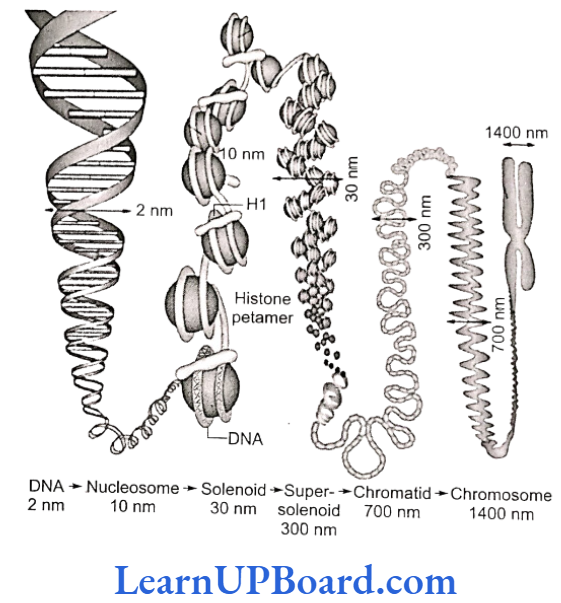
- The nucleosomes further coil to form a solenoid.
- It has diameter of 30 nm as found in chromatin.
- The beads-on-string structure in chromatin is packaged to form chromatin fibers that are further coiled and condensed at the metaphase stage of cell division to form chromosomes.
- Packaging at higher level requires additional set of proteins (acidic). These are collectively referred to as non-histone chromosomal (NHC) proteins.
- NHC proteins are of three types:
- Scaffold or structural NHC protein
- Functional NHC protein, e.g., DNA polymerase and RNA polymerase
- Regulatory NHC protein, e.g., HMG (high mobil- ity group) proteins that control gene expression)
- In a typical nucleus, some regions of chromatin are loosely packed (and stain light) and are referred to as euchromatin.
- The chromatin that is more densely packed and stains dark is referred to as heterochromatin. Specifically, euchromatin is said to be transcriptionally active while heterochromatin is said to be transcriptionally inactive.
Chemical Composition of Chromosome
A chromosome consists of the following chemical compositions:
- DNA: 40%
- RNA: 1.2%
- Histone protein: 50%
- Acidic proteins: 8.5%
- Lipid: Traces
- Ca, Mg2, Fe12: Traces
NEET Biology Notes Molecular Basis Of Inheritance Search For Genetic Material
The following experiments prove that DNA is the genetic material.
Evidence from Bacterial Transformation
- The transformation experiments conducted by Frederick Griffith in 1928 are of great importance in establishing the nature of genetic material.
- He used two strains of bacterium Diplococcus or Streptococcus pneumoniae or Pneumococcus, i.e., S-3 and R-2.
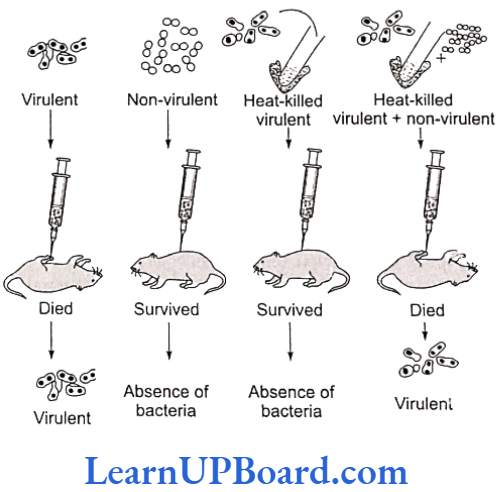
- Smooth (S) or capsulated type: These have a mucous coat and produce shiny colonies. These bacteria are virulent and cause pneumonia.
- Rough (R) or non-capsulated type: Mucous coat is absent and these produce rough colonies. These bacteria are non-virulent and do not cause pneumonia.
The experiment can be described in the following four steps: - Smooth-type bacteria were injected into mice. The mice died as a result of pneumonia caused by bacteria.
S strain Injected into mice → Mice died ii. - Rough-type bacteria were injected into mice. The mice lived and pneumonia did not occur.
R strain Injected into mice →→ Mice lived - Smooth-type bacteria, which normally cause disease, were heat-killed and then injected into mice. The mice lived and pneumonia was not caused.
S strain (heat-killed) → Injected into mice → Mice lived - Rough-type bacteria (living) and smooth- type heat-killed bacteria (both known not to cause disease) were injected together into mice. The mice died due to pneumonia and virulent smooth-type living bacteria could also be recovered from their dead bodies.
S strain (heat-killed) + R strain (living) →Injected into mice → Mice died
- From the fourth step of the experiment, he concluded that some rough-type bacteria (non-virulent) were transformed into smooth-type bacteria (virulent).
- This occurred perhaps due to the absorption of some transforming substance by rough-type bacteria from heat-killed smooth-type bacteria.
- This transforming substance from smooth-type bacteria caused the synthesis of capsule which resulted in the production of pneumonia and the death of mice.
- Therefore, transforming principle appears to control genetic characters (e.g., capsule, as in this case). However, the biochemical nature of genetic material was not defined from his experiments.
Biochemical Characterization of Transforming Principle
- Later, Avery, Macleod, and McCarty (1944) repeated the experiment in vitro to identify the biochemical nature of transforming substance. They proved that this substance is DNA.
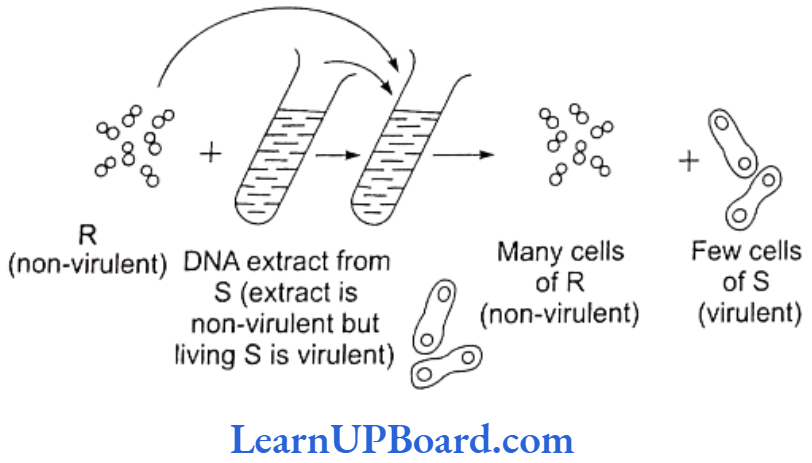
- Pneumococcus bacteria cause disease when capsule is present. Capsule production is under genetic control.
- In the experiment, rough-type bacteria (non-capsulated and non-virulent) were grown in a culture medium to which DNA extract from smooth-type bacteria (capsu- lated and virulent) was added.
- Later, the culture showed the presence of smooth-type bacteria also in addition to rough type.
- This is possible only if the DNA of smooth-type bacteria was absorbed by the rough-type bacteria which developed capsule and became virulent.
- This process of transfer of characters of one bacterium to another by taking up DNA from solution is called transformation.
- When DNA extract was treated with DNase (an enzyme that destroys DNA), transformation did not occur.
- Transformation occurred when proteases and RNases were used. This clearly shows that DNA is the genetic material.
Evidence from Experiments with Bacteriophage
- T2 bacteriophage is a virus that infects bacterium E. coli and multiplies inside it.
- T2 phage is made up of DNA and protein coat.
- Thus, it is the most suitable material to determine whether DNA or protein contains information for the production of new virus (phage) particles.
- Hershey and Chase (1952) demonstrated that only the DNA of the phage enters the bacterial cell and, therefore, contains necessary genetic information for the assembly of new phage particle.
- The functions of DNA and proteins could be found out by labeling them with radioactive tracers.
- DNA contains phosphorus but not sulfur.
- Therefore, phage DNA was labeled with p32 by grow- ing bacteria infected with phages in culture medium containing 32p.
- Similarly, the protein of phage contains sulfur but no phosphorus.
- Thus, the phage protein coat was labeled with S35 by growing bacteria infected with phages in another culture medium containing 35S.
- After the formation of labeled phages, three steps were followed:
- Infection: Both types of labeled phages were al- lowed to infect normally cultured bacteria in sep- arate experiments.
- Blending: These bacterial cells were agitated in a blender to break the contact between virus and bacteria.
- Centrifugation: The virus particles were separated from the bacteria by spinning them in a centrifuge.
molecular basis of inheritance class 12 notes bank of biology
- After centrifugation, the bacterial cells showed the presence of radioactive DNA labeled with p32 while radioactive protein labeled with $35 appeared on the outside of bacteria cells (i.e., in the medium).
- Labeled DNA was also found in the next generation of phage.
- This clearly showed that only DNA enters the bacterial host and not the protein.
- DNA, therefore, is the infective part of virus and also carries all genetic information.
- This provided the unequivocal proof that DNA is the genetic material.
Properties of Genetic Material
- Following are the properties and functions which should be fulfilled by a substance if it is to qualify as genetic material.
- It should be chemically and structurally stable.
- It should be able to transmit faithfully to the next generation, as Mendelian characters.
- It should also be capable of undergoing mutations.
- It should be able to generate its own kind (replication).
- This can be concluded after examining the above written qualities. DNA is more stable and is preferred as genetic material due to the following reasons:
- Free 2’OH of RNA makes it more labile and easily degradable. Therefore, DNA in comparison is more stable.
- The presence of thymine at the place of uracil also confers additional stability to DNA.
- RNA being unstable mutates at a faster rate.
RNA World
- RNA was the first genetic material.
- There are evidences to suggest that essential life processes such as metabolism, translation, and splicing evolved around RNA.
- RNA used to act as a genetic material as well as a catalyst.
- There are some important biochemical reactions in systems that are catalyzed by RNA catalysts and not by proteinaceous enzymes (e.g., splicing).
- RNA being a catalyst was reactive and, hence, unsta- ble.
- Therefore, DNA has evolved from RNA with chemical modifications that make it more stable.
- DNA being double stranded and having complementary strand further resists changes by evolving a process of repair.
- RNA is an adapter, structural molecule, and in some cases catalytic.
- Thus, RNA is a better material for the transmission of information.
NEET Biology Notes Molecular Basis Of Inheritance Replication Of DNA
- The Watson-Crick model of DNA immediately suggested that the two strands of DNA would separate.
- Each separated or parent strand serves as a template (model or guide) for the formation of a new but complementary strand.
- Thus, the new or daughter DNA molecules formed would be made of one old or parental strand and an- other newly formed complementary strand.
- This method of formation of new daughter DNA molecules is called the semi-conservative method of replication.
- The following experiment suggests that DNA replication is semi-conservative.
- Messelson and Stahl (1958) conducted experiment using heavy nitrogen (15N) to determine whether the concept of semi-conservative replication is correct.
- They used cesium chloride (CSCI) gradient centrifugation technique for this purpose.
- A dense solution of CsCl, on centrifugation, forms density-gradient bands of a solution of lower density at the top that increases gradually towards the bottom with highest density.
- If the DNAs of different densities are mixed with CsCl solution, these would separate from one another and would form a definite density band in the gradient along with CsCl solution.
- Meselson and Stahl created DNA molecules of different densities by using normal nitrogen, 14N, and its heavy isotope, ‘N.
- For this purpose, E. coli was grown in 15NH,Cl-containing culture medium for many generations, to make bacterial DNA completely heavy.
- This non-radioactive or heavy DNA (incorporating 15N) had more density than the DNA with normal nitrogen (14N).
- Bacteria were then transferred to the culture medium containing only normal nitrogen (NH,CI). The change in density was observed by taking DNA samples periodically.
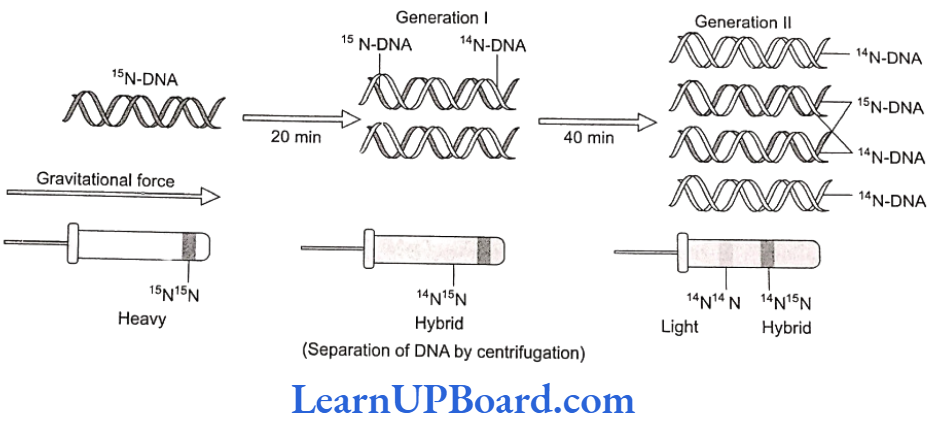
- If DNA replicates semi-conservatively, then each heavy (N) DNA strand should separate and each separated strand should acquire a light (N) partner after one round of replication.
- This should be a hybrid DNA made of two strands, i.e., 14N-15N.
- Meselson and Stahl observed that such DNA was actually half-dense indicating the presence of hybrid DNA molecules.
- After the second round of replication, there would be four DNA molecules.
- Of these, two molecules would be hybrid (14N-15N) showing half density as earlier and the remaining two molecules would be made of light strands (14N-14N). Thus, after the second generation, the same half-dense band (4N-15N) was seen but the density of light bands (14N-14N) increased.
- Meselson and Stahl’s work as such provided the confirmation of the Watson-Crick model of DNA and its semi-conservative replication.
- Taylor proved the semi-conservative mode of chromosome replication in eukaryotes using tritiated thymidine in the root of Vicia faba (faba beans).
- Cairns proved the semi-conservative mode of replication in E. coli by using tritiated thymidine (H3-tdR) in the autoradiography experiment.
- He proposed the 6-model for replication in circular DNA.
Mechanism of DNA Replication
DNA replication involves the following four major steps:
- Initiation of DNA replication
- Unwinding of helix
- Formation of primer strand
- Elongation of new strand
Initiation of Replication
- The replication of DNA always begins at a definite site called the origin of replication.
- Prokaryotes have single origin of replication.
- It is called ori-c in E.coli.
- On the other hand, eukaryotes have several thousand origins of replication.
Unwinding of Helix
- DNA replication requires that the double helical parental molecule is unwound so that its internal bases are available to the replication enzymes.
- Unwinding is brought about by enzyme helicase which is ATP-dependent.
- The unwinding of DNA molecule into two strands results in the formation of Y-shaped structure, called replication fork.
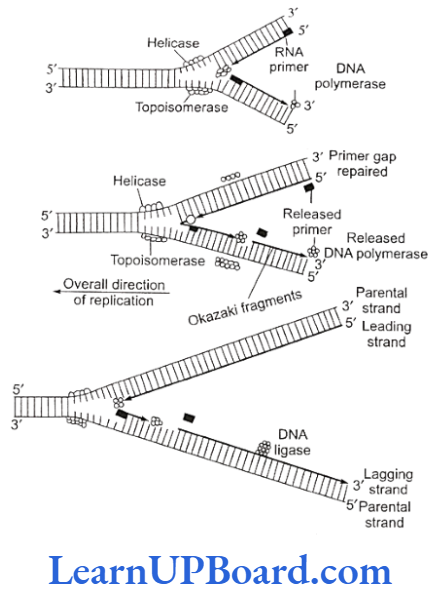
- These exposed single strands are stabilized by a protein known as single-strand binding (SSB) protein.
- Due to unwinding, a supercoiling develops on the end of DNA opposite to the replicating fork.
- This tension is released by enzyme topoisomerase.
Formation of Primer Strand
- A new strand is to be synthesized opposite to the parental strands. DNA polymerase 3 is the true replicase in E. coli. It is incapable of initiating DNA synthesis, i.e., it is unable to deposit the first nucleotide in a daughter (new) strand without the primer.)
- Another enzyme, known as primase, synthesizes a short primer strand of RNA.
- The primer strand then serves as a stepping stone (to start error-less replication).
- Once the initiation of DNA synthesis is completed, this primer RNA strand is then removed enzymatically.
Elongation of New Strand
- Once the primer strand is formed, DNA replication occurs in 5′-3′ direction, i.e., during the synthesis of a new strand, deoxyribonucleoside triphosphates (dATP, dGTP, dTTP, dCTP) are added only to the free 3’OH end.
- Thus, the nucleotide at the 3′ end is always the most recently added nucleotide to the chain.
- As DNA replication proceeds on the two parental strands, the synthesis of daughter or new strand occurs continuously along the parent 3’5′ strand.
- It is now known as the leading daughter strand.
- The synthesis of another daughter strand along the other parental strand, however, takes place in the form of short pieces.
- This is called the lagging daughter strand. These short pieces of DNA are known as the Okazaki fragments. These segments are about 1,000-2,000 nucleotides long in prokaryotes.
- Hence, DNA replication is semi-discontinuous. The discontinuous pieces of lagging strand are joined together by the enzyme DNA ligase (after the removal of primer) to form continuous daughter strand.
- Thus, two DNA molecules are now formed from one molecule.
- Each of these daughter DNA molecules is made of two strands, of which one is old (parental) and the other one is new or complementary strand.
- DNA polymerase is the most important enzyme of DNA replication.
- DNA polymerases are of three types in prokaryotes: DNA polymerase I, II, and III.
bank of biology class 12 molecular basis of inheritance
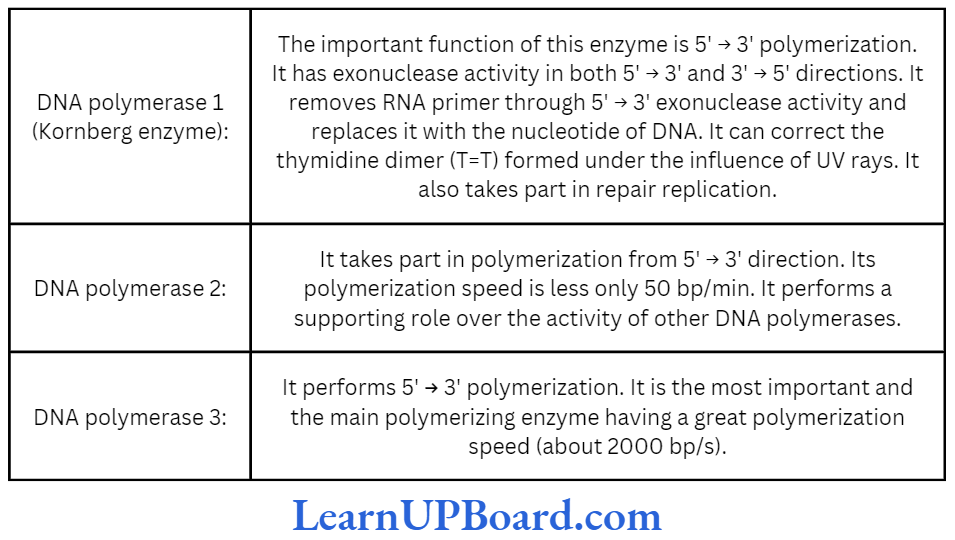
- DNA polymerases 2 and 3 have only 3′ 5′ exonuclease activity.
- Kornberg (1956) succeeded in demonstrating the in vitro synthesis of DNA molecule using a single strand of DNA as a template.
- He extracted and purified an enzyme from E. coli which was capable of linking free DNA nucleotides, in the presence of ATP as an energy source, to form complementary strand.
- He called it DNA polymerase. DNA polymerase I is called the Kornberg enzyme.
- In eukaryotes, DNA polymerases are of five types: DNA polymerase a, ẞ, 7, 8, and ɛ.
- In eukaryotes, the replication of DNA takes place at the S-phase of the cell cycle. The replication of DNA and the cell division cycle should be highly coordinated. A failure in cell division after DNA replication” results in polyploidy (a chromosomal anomaly).
NEET Biology Notes Molecular Basis Of Inheritance Structure Of RNA
RNA is present in all living cells. It is laevo rotatory and is responsible for learning and memory. It is found in the cytoplasm as well as the nucleus.
Types of RNA
- In bacteria, there are three major types of RNAs: mRNA (messenger RNA), tRNA (transfer RNA), and rRNA (ribosomal RNA).
- All three RNAs are needed to synthesize a protein in a cell.
- The mRNA provides the template, the tRNA brings aminoacids and reads the genetic code, and the rRNAs play structural and catalytic role during translation.
- RNA is generally involved in protein synthesis but in majority of plant viruses, it serves as a genetic material. Therefore, there are two major types of RNAs: genetic RNA and non-genetic RNA.
Genetic RNA
Fraenkel-Conrat showed that RNA present in TMV (tobacco mosaic virus) is a genetic material. Since then, it is established that RNA acts as a genetic material in most plant viruses.
Non-Genetic RNA
Non-genetic RNA is commonly present in cells where DNA is the genetic material. It is synthesized on DNA template. It is of the following three major types:
- mRNA
- It was reported by Jacob and Monad.
- It carries genetic information present in DNA and, so, is called working copy.
- It constitutes about 3-5% of the total RNA pre- sent in the cell.
- The molecular weight varies from 25,000 to 1,00,000.
- It is about 300 nucleotide long at minimum.
- Structure of prokaryotic mRNA: It is polycistronic in nature, i.e., several cistrons (functional part of DNA) form a single mRNA. Thus, each mRNA has a message to produce several polypeptides. Average life is 2 min.
At the 5′-end near initiation codon, a sequence of fixed bases is found. It is called the Shine Dal- garno (SD) sequence (5’AGGAGGU3′). It helps in the correct binding of ribosomal subunit (30 S) on it.
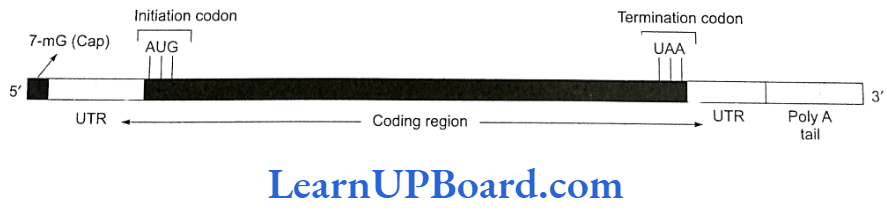
- Structure of eukaryotic mRNA: It is monocistronic in nature and has a message to produce on polypeptide only. It is metabolically stable and its life varies from a few hours to days.
- UTR (untranslated region): These are sequences of RNA before the start or initiation codon and after the stop or termination codon. These are not translated and are transcribed as part of the same transcript as the coding region. Such UTRS pro- vide stability to mRNA and also increase translational efficiency.
- rRNA
- rRNA was reported by Kuntz. It is the most stable type of RNA and is a constituent of ribosomes. It forms about 80% of the total cellular RNA.
- In eukaryotes, four types of rRNAs are found: 28 S, 18 S, 5.85 S, and 5 S, whereas in prokaryotes, three types of rRNAs are found: 23 S, 16 S, and 5 S. These are synthesized by genes present on the DNA of several chromosomes found within a region known as nucleolar organizer.
- tRNA
- The existence of tRNA was postulated by Crick. It is also known as soluble RNA (SRNA). tRNAs are the smallest molecules that carry amino acids to the site of protein synthesis. These constitute about 15% of the total cellular RNA.
- tRNA acts as an intermediate molecule between the triplet code of mRNA and the amino acid se- quence of polypeptide chain. All tRNAs have almost the same basic structure. There are over 60 types of tRNAs.
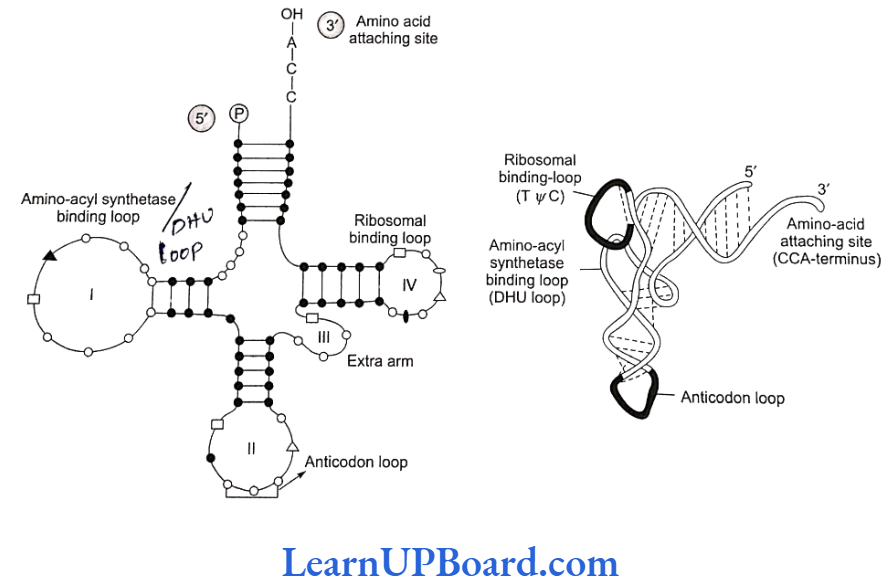
- Structure of tRNA: The cloverleaf model of tRNA is a two-dimensional model suggested by Holley et.al. A tRNA molecule appears like a cloverleaf, being folded with three or more double helical regions (stem), having loops also. The three-dimensional structure of tRNA was proposed to be inverted L-shaped by Kim and Klug.
- All tRNA molecules commonly have a guanine residue at their 5′ terminal end. At their 3′ end, un- paired CCA sequence is present. Amino acid gets attached at this end only. The number of nucleo- tides varies from 77 (tRNA alanine) to 207 (tRNA1y- rosine).
- There are three loops in tRNA.
- Amino acyl synthetase binding loop, also called DHU loop.
- Ribosomal binding loop with seven unpaired bases. It is also called TVC loop.
- Anti-codon loop with seven unpaired bases. Out of the seven bases in the anti-codon loop, three bases act as anti-codon for a particular triplet codon present on mRNA.
NEET Biology Notes Molecular Basis Of Inheritance Gene Expression
- DNA, being the genetic material, carries all the informations necessary to program the functions of a cell by controlling the synthesis of enzymes or proteins.
- Beadle and Tatum put forward a theory-one gene one enzyme-in support of the earlier hypothesis that enzymes are proteinaceous in nature and each is produced by a single gene.
- They conducted experiments on the nutritional strains of pink mold, Neurospora crassa.
- This fungus grows on simple nutrient medium and has the ability to synthesize all its cellular components. Such an organism is called prototroph.
- An organism that is unable to synthesize a particular cellular metabolite such as an amino acid or a coenzyme is called auxotroph.
- Beadle and Tatum produced arginine (an amino acid) auxotrophs (mutants of Neurospora unable to synthesize arginine) by giving X-rays treatment to the cells. Arginine synthesis passes through the following path.
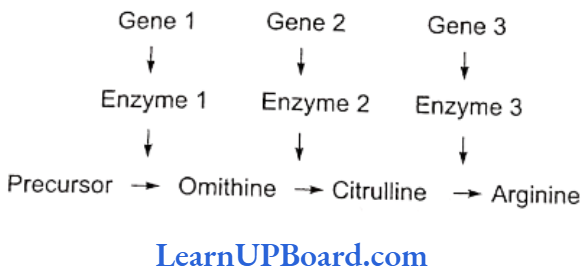
- They found that any step of this metabolic chain could be blocked by a mutation in a specific enzyme catalyzing the reaction, each enzyme representing a different gene product.
- Thus, Beadle, and Tatum reached a conclusion that each gene functions to produce a single enzyme.
- Some proteins, e.g., hemoglobin and other quaternary proteins, are made up of two or more than two poly- peptide chains. After this, the “one gene one enzyme” theory was modified into “one gene one polypeptide” hypothesis by Yanofsky. Later, Jacobson and Balti- more proposed “one mRNA one polypeptide” hypothesis.
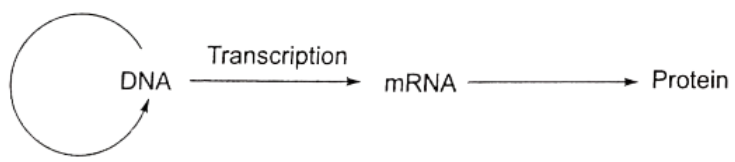
- Gene and protein: A gene expresses itself by protein synthesis.
- When a particular gene is expressed (i.e., controls a function or a reaction), its information is copied into another nucleic acid, i.e., mRNA, which in turn directs the synthesis of specific proteins.
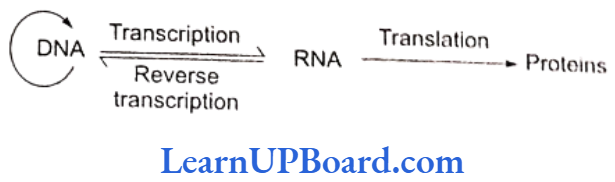
- These concepts form the central dogma of molecular biology. This has been shown by F. Crick in.
- An exception to this one-way flow of information was reported in 1970.
- H. Temin and D. Baltimore independently discovered reverse transcription in some viruses.
- These viruses can code an enzyme, reverse transcriptase, which can code DNA on RNA template.
- This discovery was important in understanding cancer and, hence, these two scientists were awarded Nobel Prize.
- Commoner suggests circular flow of information.
NEET Biology Notes Molecular Basis Of Inheritance Mechanism Of Protein Synthesis
The process of protein synthesis consists of two major steps:
- Transcription or synthesis of mRNA on DNA
- Translation or synthesis of proteins along mRNA
Transcription
- The transfer of genetic information from DNA to mRNA is known as transcription. The segment of DNA that takes part in transcription is called transcription unit. It has three components.
- A promoter
- The structural gene
- A terminator
- Promoter sequences are present upstream (5′ end) of the structural genes of a transcription unit.
- The binding sites for RNA polymerase lie within the promoter sequence.
- Certain short sequences within the promoter sites are conserved. In prokaryotes, at 10 bp upstream from the start point lies a conserved sequence described as -10 sequence TATAAT or the “Pribnow box” and -35 sequence TTGACA or the “recognition sequence.”
molecular basis of inheritance pyq neet
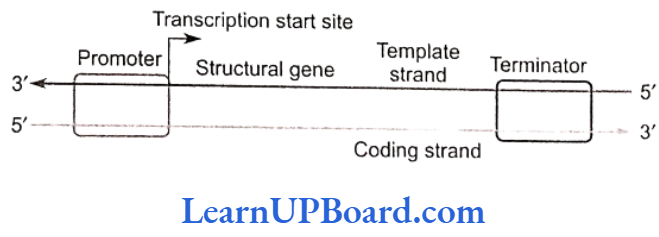
- Structural gene is a part of that DNA strand which has 3′ 5′ polarity, as transcription occurs in 5′ → 3′ direction. This strand of DNA that directs the synthesis of mRNA is called the template strand. The complementary strand is called non-template or coding strand. It is identical in base sequence to RNA transcribed from the gene, only with U in place of T.
- Terminator is present at the 3′ end of coding strand and defines the end of the process of transcription.
Mechanism of Transcription
- RNA polymerase binds to the promoter region of DNA and the process of transcription begins.
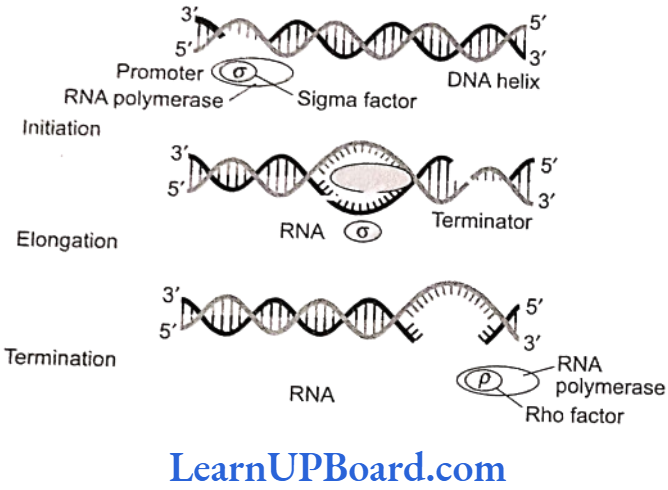
- RNA polymerase moves along the DNA helix and unwinds it.
- One of the two strands of DNA serves as a template for RNA synthesis.
- This results in the formation of complementary RNA strand.
- It is formed at a rate of about 40 to 50 bp/s.
- RNA synthesis comes to a stop when RNA polymerase reaches the terminator sequence.
- The transcription enzyme, i.e., RNA polymerase, is only of one type in prokaryotes and can transcribe all types of RNAs.
- RNA polymerase is a holoenzyme that is represented as (αBB’w)σ.
- The molecular weight of holoenzyme is 4,50,000.
- The enzyme without σ subunit is referred to as core enzyme.
- Though the core enzyme is capable of transcribing DNA into RNA, transcription starts non-specifically at any base on DNA.
- It is σ subunit which confers specificity. Rho factor (p) is required for the termination of transcription.
- Sigma factor (o): Binds to the promoter site of DNA and initiates transcription.
- Core complex: It continues the transcription.
- Rho factor (p): It terminates the transcription; its molecular weight is 55,000.
Transcription in Eukaryotes
- In eukaryotes, the promoter site is recognized by the presence of specific nucleotide sequence called TATA box or the Hogness box (7 base pair long-TATATAT or TATAAAT) located 20 bp upstream to the start point.
- Another sequence is the CAAT box present between -70 bp and 80 bp.
- In eukaryotes, the RNA polymerases are of three types: RNA polymerase I, RNA polymerase II, and RNA polymerase III.
- The functions of different RNA polymerases in eukaryotes are as follows:
- RNA polymerase 1-5.8 S, 18 S, and 28 S rRNA synthesis.
- RNA polymerase 2-hnRNA (heterogeneous nuclear RNA) and mRNA synthesis.
- RNA polymerase 3-tRNA, scRNA (small cytoplasmic RNA), 5S rRNA, and snRNA (small nuclear RNA) synthesis.
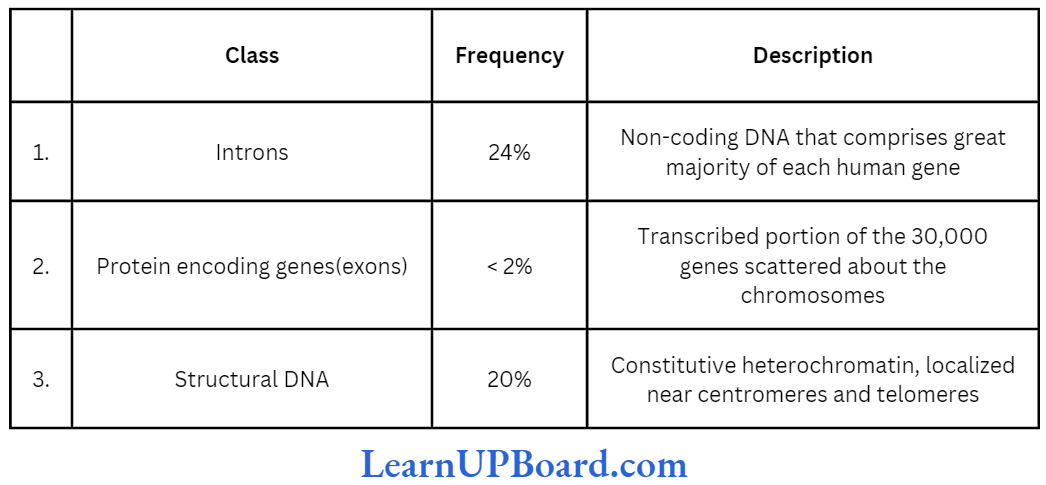
- The gene in eukaryotes, however, is made of several pieces of base sequence coding for amino acids called exonic DNA, separated by stretches of non-coding sequences, commonly called intronic DNA.
- Thus, the information on the eukaryotic gene for assembling a protein is not continuous but split.
- This discovery is the result of works of Richard J. Roberts and Philip Sharp.
- They shared 1993 Nobel Prize for Physiology and Medicine for the discovery of “split genes” in higher organisms.
- Most eukaryotic genes contain very long base sequences, all of which do not necessarily form mature mRNA.
- The coding DNA sequences of the gene are called exons and the intervening non-coding DNA sequences are called introns.
- All introns have GU at the 5′ end and AG at the 3′ end. Depending on the size of gene, the number and length of exons may vary from a few to more than 50 nucleotides. Exons alternate with stretches of DNA that contain no genetic information introns.
- The nascent RNA synthesized by RNA polymerase II is called hnRNA.
- It contains both unwanted base sequences (transcribed from introns) alternated with useful base sequences (transcribed from exons).
- This primary transcript is converted into functional mRNA after post-transcriptional processing which involves three steps:
- Modification of 5′ end by capping: Capping at the 5′ end occurs rapidly after the start of transcription.
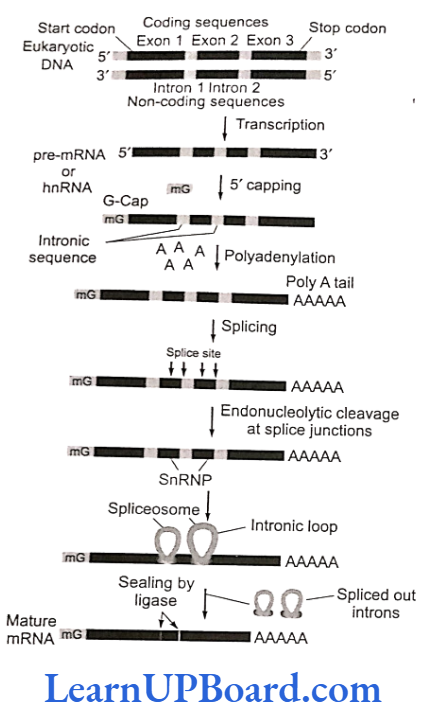
- The guanosine methylated at the seventh position is added at 5′ with the help of enzyme guanyl transferase.
- Cap is essential for the formation of mRNA-ribosome complex.
- Translation is not possible if cap is lacking, be- cause cap is identified by 18S rRNA of ribosome unit.
- Polyadenylation at 3′ end (tailing): Poly (A) is added to the 3′ end of newly formed hnRNA with the help of enzyme poly A polymerase. It adds about 200-300 adenylate residues.
- Splicing of hnRNA (tailoring): In eukaryotes, the coding sequences of RNA (exons) are interrupted by non-coding sequences (introns).
- snRNA and protein complex called small nuclear ribonucleoprotein (or snRNPs or snurps) play important role in this process.
- Here, the introns are removed and the exons come in one plane.
- This process is called splicing through which a mature mRNA is produced.
- Normally, mRNA carries the codons of a single complete protein molecule (monocistronic mRNA) in cukaryotes, but in prokaryotes, it carries codons from several adjacent DNA cistrons and becomes much longer in size (polycistronic mRNA).
Genetic Code
- The term genetic code was coined by Gamow.
- DNA (or RNA) carries all the genetic information.
- It is expressed in the form of proteins.
- Proteins are made up of 20 different types of essential amino acids.
- The information about the number and sequence of these amino acids forming proteins is present in DNA and is passed on to mRNA during transcription.
- Genetic code is an mRNA sequence containing coded information for one amino acid. It consists of three nu- cleotides (triplet).
- Thus, for 20 amino acids, 64 (4 x 4 x 4 or 43 = 64) minimum possible permutations are available.
- This important discovery was the result of experiments by Marshall W. Nirenberg and J. Heinrich Matthaei and later by H.G. Khorana. Nirenberg and Khorana shared the 1968 Nobel Prize with R.W. Holley who gave the details of tRNA structure. Nirenberg and Mathaei used a synthetic poly(U) RNA and deciphered the code by translating this as polyphenylalanine. Hargobind Kho- rana, using synthetic DNA, prepared polynucleotide with known repeating sequence, CUCUCUCUCUCU; it produced only two amino acids-leucine (CUC) and serine (UCU).
Properties of Genetic Code
- Triplet code: Three adjacent nitrogen bases constitute a codon which specifies the placement of one amino acid in a polypeptide.
- Start signal: Polypeptide synthesis is signaled by two initiation codons-AUG or, rarely, GUG. The first or initiating amino acid is methionine. The initiating codon on mRNA for methionine is AUG. Initiating methionine occurs in formylated state in prokaryotes. At other positions, methionine is non-formylated. Both these methionines are carried by different tRNAs. Rarely, GUG also serves as initiation codon. It normally codes for valine but if present at the initiating position, it would code for methionine. So, GUG is an ambiguous codon.
- Stop signal: Polypeptide chain termination is signaled by three termination codons-UAA (ochre), UAG (amber), and UGA (opal). They do not specify any amino acid and were, hence, called nonsense codons. Whenever present in mRNA, these bring about termination of polypeptide chain and, thus, act as stop signals. Codons UAA, UAG, UGA, AUG, and GUG are also called punctuation codons.
- Commaless: The genetic code is continuous and does not possess pauses (meaningless base) after each tri- plet. If a nucleotide is deleted or added, the whole genetic code will read differently. Thus, a polypeptide having 50 amino acids shall be specified by a linear sequence of 150 nucleotides. If a nucleotide is added or deleted in the middle of this sequence, the amino acids before this will be the same but subsequent amino acids will be quite different.
- Universal code: The genetic code is applicable universally, i.e., a codon specifies the same amino acid from a virus to a human being or a tree.
- Non-overlapping code. Each codon is independent and one codon does not overlap with the next one.
- Degeneracy of code: Since there are 64 triplet codons and only 20 amino acids, the incorporation of some amino acids must be influenced by more than one codon. Only tryptophan and methionine are specified by single codons. All other amino acids are specified by 2-6 codons. The latter are called degenerate codons.
Wobble Hypothesis
- A change in nitrogen base at the third position of a codon does not normally cause any change in the expression of the codon because the codon is mostly read by the first two nitrogen bases (wobble hypothesis of Crick).
- The mutation that does not cause any change in the expression of the gene is called silent mutation.
- The position of the third nitrogen base in a codon which does not influence the reading of the codon is termed as wobble position.
- It pairs with the first position in anticodon. It means that the specificity of codon is determined by the first two bases.
- Wobbling helps one tRNA to read more than one codon and, thus, provides economy in the number of tRNA molecules at the time of translation.
Mutations and Genetic Code
- In a hypothetical mRNA, for example, the codons would normally be translated as follows.

- The insertion of single base G between the third and fourth bases produces a completely different protein from the earlier one.

- Similarly, the deletion of single base C at the fourth place produces a new chain of amino acids and, hence, a different protein.

- The insertion or deletion of one or two nitrogenous bases changes the reading frame from the point of insertion or deletion.
- Such mutations are called frame shift mutations.
- However, the insertion or deletion of three or its mul- tiple bases leads to the insertion or deletion of one or multiple codons. Hence, one or multiple amino acids and reading frames remain unaltered from that point onwards.
- This forms the genetic basis of proof that codon is a triplet and that it is read in a contiguous manner.
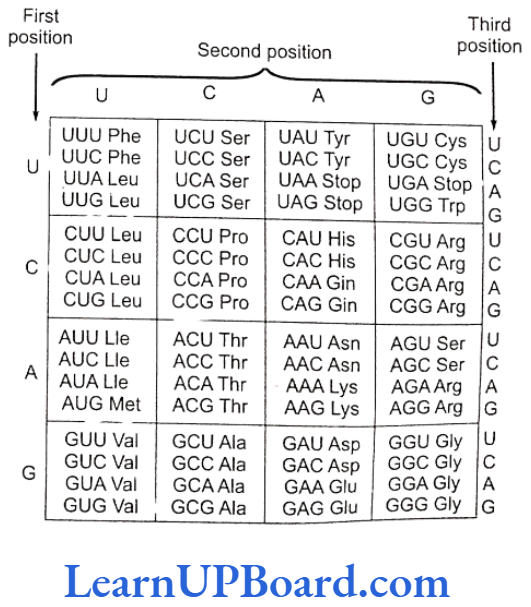
Translation
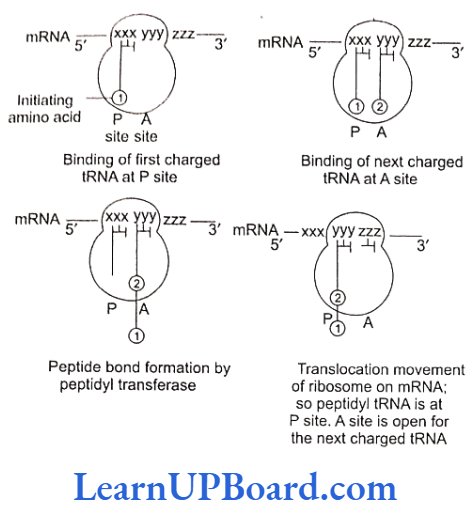
- Translation is the mechanism by which triplet base sequences of mRNA molecules are converted into a specific sequence of amino acids in a polypeptide chain. It occurs on ribosomes.
- The major steps are as follows:
- Activation of amino acids: In the presence of enzyme aminoacyl-tRNA synthetase (E), specific amino acids (AA) bind with ATP.
- Charging of tRNA: The AA, -AMP-E, com- plex formed in the first step reacts with a specific tRNA. Thus, amino acid is transferred to tRNA. As a result, the enzyme and AMP are liberated.
- Formation of polypeptide chain: It is completed in three steps.
- Chain initiation: It requires three initiation factors in prokaryotes-IF3, IF2, and IF.
Nine initiation factors are required in eukaryotes. These are elF2, eIF3, eIF1, eIF4A, eIF4B eIF4c, eIF4D, eIFs, and eIF6.- Binding of mRNA with smaller subunit of ribosomes (30S/40S): IF, is involved in prokaryotes while eIF2 is involved in eukaryotes. It involves interaction be- tween the Shine Delgarno sequence and the 3′ end of 16S rRNA in prokaryotes.
- Binding of 30S/40S-mRNA complex with tRNA: Non-formylated methionine is attached with tRNA in eukaryotes and formylated methionine in prokaryotes. IF2 is involved in prokaryotes while eIF3 is involved in eukaryotes.
- Attachment of larger subunit of ribosomes: Initiation factors in eukaryotes are eIF, and eIF4. Initiation factor in prokaryotes is IF.
- Chain elongation
- Ribosomes have two sites for binding amino acyl tRNA: (1) amino acyl or A site (acceptor site) and (2) peptidyl site or P site (donor site).
- A second charged tRNA molecule along with its appropriate amino acid approaches the ribosome at the A site close to the P site.
- Its anticodon binds to the complementary codon of mRNA chain.
- A peptide bond is formed between the COOH group of the first amino acid (methionine) and the NH2 group of the second amino acid.
- The formation of peptide bond requires energy and is catalyzed by enzyme peptidyl transferase. The initiating formyl methionine or methionine tRNA can bind only with the P site.
- All other newly coming aminoacyl tRNA bind to the A site.
- The elongation factors required for prokaryotes are EF-Tu and EF-Ts and that required for eukaryotes is eEF.
- Translocation is the movement of ribo- some on mRNA. It requires EF-G in prokaryotes and eEF, in eukaryotes.
- Chain initiation: It requires three initiation factors in prokaryotes-IF3, IF2, and IF.
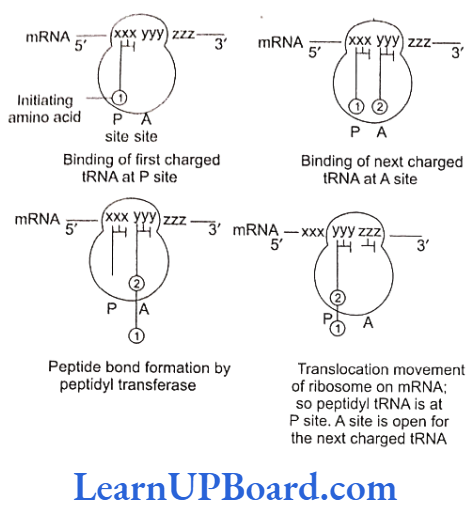
- Chain termination: The termination of polypeptide is signalled by one of the three terminal codons in the mRNA. The three terminal codons are UAG (amber), UAA (ochre), and UGA (opal).
- AGTP dependent factor known as release factor is associated with the termination codon. It is eRF1 in eukaryotes and RF1 ND RF2 in projaryotes.
- At the time of termination, the terminal codon immediately follows the last amino acid codon. After this, the polypeptide chain, tRNA, and mRNA are released and the sumunits of ribosomes get dissociated.
- Gene expression is the mechanism at the molecular level by which a gene is able to express itself in the phenotype of an organism.
- In the translation unit, mRNA has some sequences that are not translated and are referred to as untranslated regions (UTR).
- The UTRs are present at both 5′ end (before the start codon) and 3′ end (after the stop codon).
- They are required for efficient translation process.
- Chain termination: The termination of polypeptide is signalled by one of the three terminal codons in the mRNA. The three terminal codons are UAG (amber), UAA (ochre), and UGA (opal).
NEET Biology Notes Molecular Basis Of Inheritance Regulation Of Gene Expression
- Constitutive genes are those genes that are constantly expressing themselves in a cell because their products are required for normal cellular activities. For example, genes for glycolysis and ATPase.
- Non-constitutive genes are not always expressing themselves in a cell. These are called luxury genes. These are switched on or off according to the requirement of cellular activities. For example, genes for nitrate reductase in plants and lactose system in E. coli. This provides maximum functional efficiency to the cell. Such regulated genes, therefore, are required to be switched on and off when a particular function is to begin or stop.
- This regulation can be achieved by any one of the following two processes:
- Induction: This is a process of gene regulation where the addition of a substrate stimulates or in- duces the synthesis of enzymes needed for its own breakdown.
- Repression: In this process of gene regulation, the addition of end product stops the synthesis of enzymes needed for its formation. This phenomenon is also known as feedback repression.
Operon
- Francois Jacob and Jacques Monod (1961) proposed a model of gene regulation, known as the operon model.
- Operon is a coordinated group of genes such as structural gene, operator gene, promoter gene, and regulator gene which function or transcribe together and regulate a metabolic pathway as a unit.
Inducible Operon (Lac Operon)
- In E. coli, the breakdown of lactose requires three enzymes.
- These enzymes are synthesized together in a coordinated manner and the unit is known as lac operon.
- Since the addition of lactose itself stimulates the production of required enzymes, it is also known as inducible system.
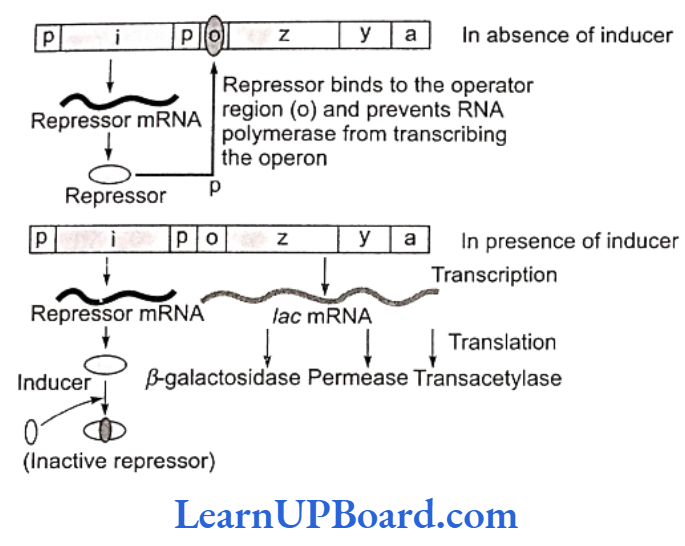
- Lac operon consists of the following genes:
- Structural genes: These genes code for the proteins needed by the cell which include enzymes or other proteins having structural functions. In lac operon, there are following three structural genes:
- lac a-Gene coding for enzyme transacetylase
- lac y Gene coding for enzyme permease
- lac z-Gene coding for enzyme ẞ-galactosidase
- Operator gene (o): It interacts with a protein molecule (the regulator molecule), which pro- motes (induces) or prevents (represses) the transcription of structural genes.
- Promoter gene (p): This gene is the recognition point where RNA polymerase remains associated.
- Regulator gene (i): This is generally known as inhibitory gene (1). This gene codes for a protein called the repressor protein. It is an allosteric protein which can exist in two forms. In one form, it is paratactic to an operator and binds with it, and in the other form, it is paratactic to the inducer (such as lactose).
- Structural genes: These genes code for the proteins needed by the cell which include enzymes or other proteins having structural functions. In lac operon, there are following three structural genes:
- The operon is switched off and on.
- The transcription or function of lac genes or structural genes depends on the operator gene.
- When the repressor protein produced by inhibitory (i) gene or regulatory gene binds to the operator (o) gene, RNA polymerase gets blocked. There is no transcription and the operon model remains in “switched off” position.
- On the other hand, when an inducer such as lactose is added, the repressor protein (produced by gene i) gets bound to it.
- The repressor, therefore, gets released from the opera- tor.
- RNA polymerase is now permitted to act.
- Hence, the transcription of lac genes now takes place and the operon model is in “switched on” position.
- All three genes are transcribed by RNA polymerase to form a single mRNA strand.
- Each gene segment of mRNA is called cistron and, therefore, the mRNA strand transcribed by more than one gene is known as polycistronic.
Tryptophan Operon-Repressible Operon System
- Operon model can also be explained using feedback repression.
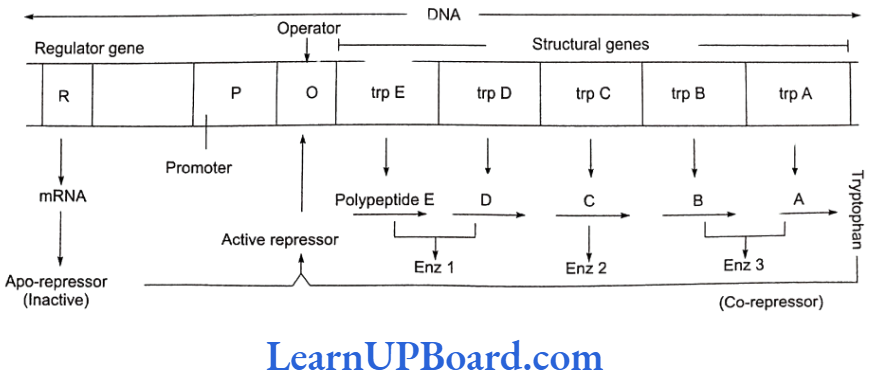
- In tryptophan (trp) operon, three enzymes are necessary for the synthesis of amino acid tryptophan.
- These enzymes are synthesized by the activity of five different genes in a coordinated manner.
- The addition of tryptophan, however, stops the produc- tion of these enzymes.
- Thus, the system is known as repressible system.
- In this system, there are five structural genes: trp A, trp B, trp C, trp D, and trp E. Three enzymes are needed for the synthesis of tryptophan-an amino acid.
- Regulatory gene (R) produces repressor protein which is known as apo-repressor because it does not get bound to the operator directly.
- Hence, the operator gene remains in “switched on” po- sition.
- Tryptophan, when added, binds to the apo-repressor and is called co-repressor.
- This apo-repressor and co-repressor complex (acti- vated repressor) now binds to the operator gene and blocks the function of RNA polymerase.
- Thus, transcription would not occur and tryptophan operon would be in “switched off” position.
- Feedback repression is functional when there is no further need of the end product and, hence, there is no requirement of continuation of this anabolic pathway.
- This operon stops the process.
- In prokaryotes, the control of the rate of transcriptional initiation is the predominant site for the control of gene expression.
Regulation of Gene Expression in Eukaryotes
- In eukaryotes, the regulation of gene expression can be exerted at four levels:
- Transcriptional level (formation of primary transcript)
- Processing level (regulation of splicing)
- Transport of mRNA from nucleus to the cytoplasm
- Translational level
- In eukaryotes, functionally related genes do not represent an operon but are present on different sites in chromosomes.
- Here, the structural gene is called split gene. It is a mosaic of exons and introns, i.e., base triplet-amino acid matching is not continuous.
- The Britten-Davidson gene battery model is most popular for eukaryotic genes.
- It proposes the occurrence of five types of genes: producer, receptor, integrator, sensor, and enhancer-silencer.
- Exons are the coding part of cistron which forms RNA.
- Introns are the non-translated part of DNA called IVS (intervening sequences) or spacer DNA.
- An intron begins with GU and ends up with an AG.
- However, the entire split gene is transcribed to form a continuous strip of hnRNA.
- This removal of non-coding intronic part and the fusion of exonic coding parts of RNA is called RNA splicing. About 50%-90% of primary transcribed RNA is discarded during processing.
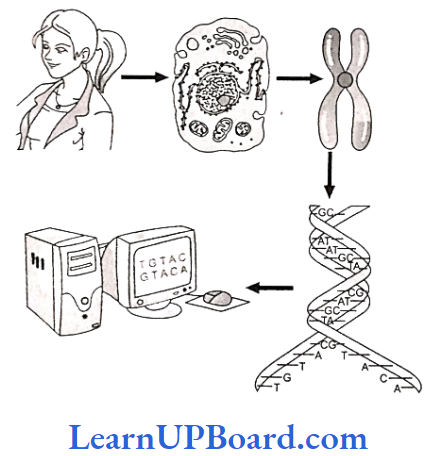
DNA Fingerprinting
- The chemical structure of everyone’s DNA is the same.
- The only difference between people (or any animal) is the order of the base pairs.
- There are so many millions of base pairs in each person’s DNA that every person has a different sequence.
- Using these sequences, everyone can be identified solely by the sequence of their base pairs.
- However, there are so many millions of base pairs due to which the task would be very time consuming.
- Instead, scientists are able to use a shorter method, because of repeating base patterns in DNA (satellite DNA).
- These patterns do not, however, give an individual “fingerprint.” But these are able to determine whether two DNA samples are from the same person, related people, or non-related individuals.
NEET Biology Notes Molecular Basis Of Inheritance VNTRS, RFLP, SSR, AND RAPD
- Each strand of DNA has stretches that contain genetic information responsible for an organism’s development (exons) and stretches that, apparently, supply no relevant genetic information at all (introns).
- Although the introns may seem useless, it has been found that they contain repeated sequences of base pairs.
- These sequences are called variable number tandem repeats (VNTRS).
- VNTRS, also called minisatellites, were discovered by Alec Jeffreys et. al. of UK.
- These consist of hypervariable repeat regions of DNA having a basic repeat sequence of 11-60 bp and flanked on both sides by restriction sites.
- The number of repeats shows a very high degree of polymophism and the size of VNTR varies from 0.1 kb to 20 kb. The length of minisatellites and the position of restriction sites is different for each person.
- Therefore, when the genomes of two people are cut using the same restriction enzyme, the length and number of fragments obtained are different for both.
- This is called restriction fragment length polymorphism (RFLP).
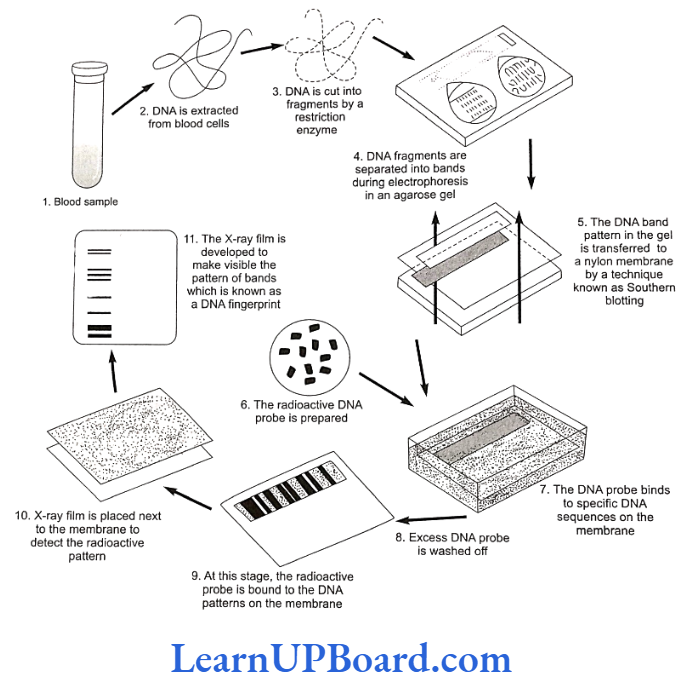
- These fragments, when separated by gel electrophoresis, and obtained on a Southern blot, constitute what is called DNA fingerprint.
- The father of DNA fingerprinting is Alec Jeffreys while the Indian experts Lalji Singh and V.K. Kashyap are known as the father of Indian technique.
Methodology of DNA Fingerprinting
The Southern blot is one way to analyze the genetic patterns which appear in a person’s DNA. It was devised by E. M. Southern (1975) for separating DNA fragments.
- Isolating the desired DNA: It can be done either chemically (by using a detergent to wash the ex- tra material from the DNA) or mechanically (by applying a large amount of pressure in order to “squeeze out” the DNA).
- Cutting the DNA into several pieces of different size: This is done using one or more restriction enzymes.
- Sorting the DNA pieces by size: The process by which size separation is done is called gel electrophoresis. The DNA is poured into a gel, such as agarose, and an electric charge is applied to the gel, with the positive charge at the bottom and the negative charge at the top. Because DNA has a slightly negative charge, the pieces of DNA will be attracted towards the bottom of the gel. The smaller pieces, however, will be able to move more quickly and, thus, further towards the bottom than the larger pieces. The different-sized pieces of DNA will, therefore, be separated by size, with the smaller pieces towards the bottom and the larger pieces towards the top.
- Denaturing the DNA fragments, so that all of the DNA is rendered single-stranded: This can be done either by heating by chemically treating the DNA in the gel using alkali.
- Blotting the DNA: The gel with size-fractionated DNA is applied to a sheet of nitrocellulose paper or nylon membrane, and then baked to permanently attach the DNA to the sheet. The Southern blot is now ready to be analyzed.
In order to analyze a Southern blot, a radioactive genetic probe is used in a hybridization reaction with the DNA in ques- tion. If an X ray is taken of the Southern blot after a radioactive probe has been allowed to bind with the denatured DNA on the paper, only the areas where the radioactive probe binds will show themselves on the film (autoradiography). This allows researchers to identify, in a particular person’s DNA, the occurrence and frequency of the particular genetic pattern contained in the probe.
Practical Applications of DNA Fingerprinting
- Solving cases of disputed paternity and maternity: Because a person inherits his or her VNTRS from his or her parents, VNTR patterns can be used to establish paternity and maternity.
- Criminal identification and forensics: DNA isolated from blood, hair, skin cells, or other genetic evidence left at the scene of crime can be compared, through VNTR patterns, with the DNA of a criminal suspect to determine guilt or innocence.
- Personal identification: The notion of using DNA fingerprints as a sort of genetic bar code to identify individuals has been discussed, but this is not likely to happen anytime in the near future. The technology required to isolate, keep on file, and then analyze millions of very specified VNTR patterns is both expensive and impractical.
NEET Biology Notes Molecular Basis Of Inheritance Genomics
- The application of DNA sequencing and genome mapping to the study, design, and manufacture of biologically important molecules is known as genomics.
- It is comparatively a more recent branch in the field of biology.
- The term genomics was introduced by Thomas Roder- ick.
- In genomics, the function of gene is identified by using the technique of reverse genetics.
- The study of genomics may be classified into three classes:
- Structural genomics: It involves the mapping and sequencing of genes.
- Functional genomics: It involves the identification of function of a particular gene.
- Application genomics: It involves the use of genomics information for crop improvement, etc.
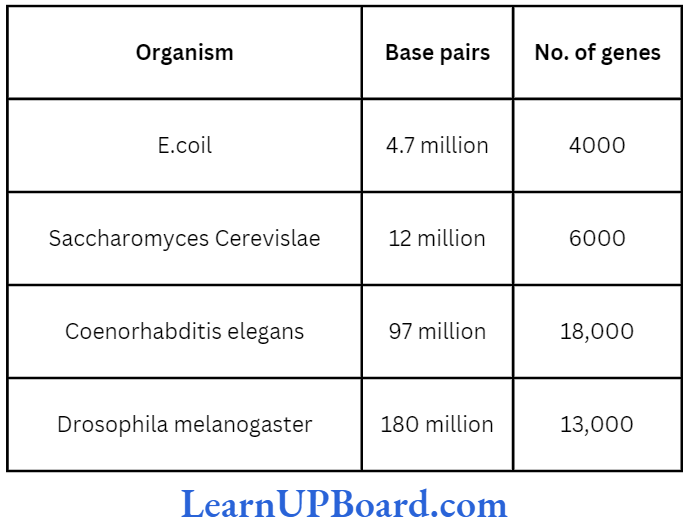
- The complete genomics of Arabidopsis and rice have been worked out. These have 130 million bp and 430 million bp, respectively.
Human Genome Project
- The Human Genome Project (HGP) was the international, collaborative research program whose goal was the complete mapping and understanding of all genes of human beings.
- All genes together are known as genome.
- The HGP as “mega project” was a 13 year project co- ordinated by the US Department of Energy and the Na- tional Institute of Health.
- An International HGP was launched in the year 1990 and completed in the year 2003.
- The International Human Genome Sequencing Consortium published the first draft of the human genome in the journal Nature in February 2001.
- Human genome is said to have approximately 3 × 10 bp, and the cost of sequencing required is US$ 3 per bp. The total estimated cost of the project would be approximately US$ 9 billion. In human ge- nome, 20,000 to 25,000 genes are present; out of them, the smallest gene is testis-determining fac- tor (TDF) gene with 14 bp and the largest gene is Duchenne muscular dystrophy gene with 2400 × 103 bp.
Goals of HGP
Following are the important goals of the HGP:
- Identification of all, approximately, 20,000-25,000 genes in human DNA.
- To determine the sequence of 3 billion chemical base pairs that make up human DNA.
- To store this information in databases.
- To improve tools for data analysis.
- To transfer related technologies to other sectors, such as industries.
- Bioinformatics, i.e., close association of HGP with the rapid development of a new area in biology.
- Sequencing of model organisms-Complete genome sequence of E.coli, S. cerevisiae, C. elegans, D. melanogaster, D. pseudoobscura, Oryza sativa, and Arabidopsis was achieved in April, 2003.
- Address the ethical, legal, and social issues (ELSI) that may arise from the project.
Methodologies
The HGP techniques include the following:
- Sequence tagged site (STS): It is a short DNA segment that occurs only once in a genome and whose ex- act location and order of bases are known. STSS serve as landmarks on the physical map of a genome. These are also called expressed sequence tags (ESTs). Genes that are expressed as RNA are referred to as ESTS.
- Sequencing the whole set of genome that contained all the coding and non-coding sequences and later assigning different regions in the sequence with functions is known as sequence annotation.
- The employment of restriction fragment length polymorphism (RFLP).
- Yeast artificial chromosomes (YACs).
- Bacterial artificial chromosomes (BACs).
- Polymerase chain reaction (PCR).
- Electrophoresis
- For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes (recall DNA is a very long polymer, and there are technical limitations in sequencing very long pieces DNA) and cloned in suitable host using specialized vectors.
- Cloning results in the amplification of each piece of DNA fragment so that it can be subsequently sequenced with ease.
- The commonly used hosts were bacteria and yeast, and the vectors were called as BAC and YAC, respectively.
- The fragments were sequenced using automated DNA sequencers that worked on the principle of the method developed by Frederick Sanger. These sequences were then arranged based on some overlapping regions present in them. This required the generation of overlapping fragments for sequencing. The alignment of these sequences was humanly not possible. Therefore, specialized computer-based programs were developed. These sequences were subsequently annotated and were assigned to each chromosome. The sequence of chromosome-1 was completed only in May, 2006. (This was the last of the 24 human chromosomes-22 autosomes and X and Y–to be sequenced.)
Salient Features of Human Genome
Some salient observations drawn from the HGP are as follows:
- The human genome contains 3164.7 million nucleotide bases.
- On an average, a gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
- The total number of genes is estimated at 30,000- much lower than the previous estimates of 80,000- 1,40,000 genes. Almost all (99.9%) nucleotide bases are exactly the same in all people.
- The functions are unknown for over 50% of discov- ered genes.
- Less than 2% of the genome codes for proteins.
- Repeated sequences make up very large portion of the human genome.
- Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times. They are thought to have no direct coding functions, but they shed light on chromosome structure, dynamics, and evolution.
- Chromosome-1 has the highest number of genes (2968) while Y-chromosome has the fewest (231).
- Scientists have identified about 1.4 million locations where single-base DNA differences (SNPs or single nucleotide polymorphism, pronounced as “snips”) occur in humans. This information promises to revolutionize the process of finding chromosomal locations for disease-associated sequences and tracing human history.
Future Thrust of HGP
Francis Collins, the director of the public funded genome project, predicted the following progress in the HGP:
By 2010
- Scientists will have developed accurate predictive tests for at least a dozen common diseases so that preventive measures may be taken in advance.
- Infertility specialists will be using sophisticated techniques of pre-implantation genetic diagnosis to screen embryos for genetic disorders and designer babies.
- Medicines will be making use of gene therapy.
By 2020
- Doctors will have “designer drugs” to treat almost every disease.
- Doctors will test patient’s genetic make-up before pre- scribing drugs.
- Doctors will be able to change the genetic make-up of a living person by germ-line therapy.
- The treatment of diseases such as cancer, schizophrenia, and depression will have transformed.
By 2030
- Gene-based healthcare will have completely developed.
- Most medical researches will be carried out using computer models rather than the living tissues of animals.
- Average life span will rise up to 90 years.
The Indian Scenario
- The Indian gene center accounts for 160 species out of 2400 species in all of the 12 megagene centers.
- Though the plant genetic resources activities got in- tensified in India in the first half of the century, these received the required impetus after the creation of NBPGR in 1976 which has headquarters at IARI (In- dian Agricultural Research Institute), New Delhi, and several regional sub-centers at different agroclimatic zones such as Jodhpur (arid areas), Trichur (humid tropical zone), and Shillong (eastern sub-tropical/sub- temperate zone).
- On March 31, 1996, NBPGR held 15,4,533 accessions of various agri-horticultural crops.
- National Facility for Plant Tissue Culture Repository (NFPTCR) was established in 1986 by the department of biotechnology at NBPGR for the conservation of germplasm of vegetatively propagated plants.
NEET Biology Notes Molecular Basis Of Inheritance Summary
- Nucleotide monomers constitute a polymer called nucleic acid. It is of two types: RNA and DNA.
- While DNA is the store house of information, RNA helps in the transfer and expression of information.
- As DNA is structurally and chemically more stable, it is a better genetic material, although both DNA and RNA serve as genetic material.
- RNA was the first to evolve, and DNA was derived from it.
- Bases in two DNA strands show hydrogen bonding (A=T, G=C) and follow Chargaff’s rule, so that both the strands are complementary and DNA replication is semi-conservative.
- The segment of DNA that codes for RNA is known as gene. During transcription, one DNA strand acts as template which directs the synthesis of complementary RNA.
- In prokaryotes, transcription and translation are continuous processes. In eukaryotes, the genes are split. Exons are interrupted by introns. Introns are removed and exons are joined to produce functional RNA.
- The mRNA contains genetic code in combination of three (triplet code) to code for an amino acid. This genetic code is read by tRNA which acts as an adapter molecule.
- There is specific tRNA for each amino acid. Each tRNA binds to amino acid at one end and with codons by H-bonding at the other end.
- Translation occurs at ribosome. Here ribozyme (rRNA enzyme) acts as catalyst which helps in peptide bond formation. The process of translation has evolved around RNA, which shows that life began around RNA.
- Since transcription and translation are energetically very expensive, they are tightly regulated. For example, lac operon, which is regulated by the amount of lactose in medium, i.e., the regulation of enzyme synthesis by its substrate.
- HGP is aimed at sequencing every base in human genome. DNA fingerprinting is used for this which is based on the principle of polymorphism in DNA sequence.
NEET Biology Notes Molecular Basis Of Inheritance Assertion-Reasoning Questions
In the following questions, a statement of Assertion (A) is followed by a statement of Reason (R).
- If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark (1).
- If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
- If Assertion is true but Reason is false, then mark (3).
- If both Assertion and Reason are false, then mark (4).
Question 1. Assertion: RNA polymerase is of three types in eukaryotes for the synthesis of all types of RNAs.
Reason: RNA polymerase consists of six types of poly- peptides along with rho factor which is involved in the termination of RNA synthesis.
Answer. 3. If Assertion is true but Reason is false, then mark (3).
Question 2. Assertion: 5S rRNA and surrounding protein complex provides binding site of tRNA.
Reason: tRNA is soluble RNA with unusual bases.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 3. Assertion: Operator gene is functional when it is not blocked by repressor.
Reason: Regulator gene produces active protein only which acts on operon system in E. coli.
Answer. 3. If Assertion is true but Reason is false, then mark (3).
Question 4. Assertion: Peptidyl transfer site is contributed by larger subunit of ribosome.
Reason: The enzyme peptidyl transferase is contributed by both 23S and 16S ribosomal subunits.
Answer. 3. If Assertion is true but Reason is false, then mark (3).
Question 5. Assertion: Teminism is unidirectional flow of information.
Reason: It requires DNA dependent RNA polymerase enzyme.
Answer. 4. If both Assertion and Reason are false, then mark (4).
Question 6. Assertion: In bacterial translation mechanism, two tRNA are required by methionine.
Reason: AUG codes for methionine and it shows non- ambiguity also.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 7. Assertion: Nutritional mutant strain of pink mold is auxotroph.
Reason: It is not able to prepare its own metabolites from the raw materials obtained from outside.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark (1).
Question 8. Assertion: In DNA fingerprinting, hybridization is done with molecular probe.
Reason: Molecular probe is small DNA segment synthesized in laboratory with known sequence that recognizes complementary sequence in RNA.
Answer. 3. If Assertion is true but Reason is false, then mark (3).
Question 9. Assertion: cDNA libraries are important to scientists in human genomics.
Reason: cDNA is synthetic type of DNA generated from mRNA.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 10. Assertion: SNP (pronounced “snips”) are common in human genome.
Reason: It is minute variation that occurs at a frequency of one in every 300 bases.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark (1).
Question 11. Assertion: A single strand of mRNA is capable of forming a number of polypeptide chains.
Reason: Termination codons occur in mRNA.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 12. Assertion: Chromosomal aberrations are caused by a break in the chromosome or its chromatid.
Reason: Duplication, deficiency, transversion, and trans- locations are the causes of chromosomal aberrations.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 13. Assertion: The lac operon model is applicable to E. coli.
Reason: E. coli. lacks a definite nucleus.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 14. Assertion: Amber codon is a termination codon.
Reason: If in mRNA, a termination codon is present, protein synthesis stops abruptly whether it is completed or not.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
Question 15. Assertion: Watson and Crick provided experimental proof of the semi-conservative nature of DNA replication.
Reason: RNA polymerase binds nucleotides in replication.
Answer. 4. If both Assertion and Reason are false, then mark (4).
Question 16. Assertion: The mRNA attaches itself to the ribosome via its 3′ end.
Reason: The mRNA has nucleotide and bases of lagging sequence.
Answer. 4. If both Assertion and Reason are false, then mark (4).
Question 17. Assertion: Replication and transcription occur in the nucleus but translation occurs in the cytoplasm.
Reason: mRNA is transferred from the nucleus into the cytoplasm where ribosomes and amino acids are available for protein synthesis.
Answer. 1. If both Assertion and Reason are true and the reason is the correct explanation of the assertion, then mark (1).
Question 18. Assertion: Cancer cells are virtually immortal until the body in which they reside dies.
Reason: Cancer is caused by damage to genes regulating the cell division cycle.
Answer. 2. If both Assertion and Reason are true but the reason is not the correct explanation of the assertion, then mark (2).
